+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | wobble uridine modification / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA uridine(34) acetyltransferase activity / tRNA carboxymethyluridine synthase / elongator holoenzyme complex / protein urmylation / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / tRNA wobble uridine modification / regulation of translation / protein transport / 4 iron, 4 sulfur cluster binding / microtubule binding ...tRNA uridine(34) acetyltransferase activity / tRNA carboxymethyluridine synthase / elongator holoenzyme complex / protein urmylation / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / tRNA wobble uridine modification / regulation of translation / protein transport / 4 iron, 4 sulfur cluster binding / microtubule binding / tRNA binding / regulation of transcription by RNA polymerase II / nucleoplasm / metal ion binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D ...Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D / Lin T-Y / Abbassi N / Hammermeister A / Chramiec-Glabik A / Kosinski J / Schaffrath R / Glatt S | |||||||||
| Funding support |  Poland, European Union, 2 items Poland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Cryo-EM structure of the fully assembled Elongator complex. Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej ...Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej Chramiec-Głąbik / Anna Biela / Dominika Dobosz / Ting-Yu Lin / Nour-El-Hana Abbassi / Alexander Hammermeister / Juri Rappsilber / Jan Kosinski / Raffael Schaffrath / Sebastian Glatt /    Abstract: Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition ...Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition of chemical groups. Modifications in the tRNA anticodons are directly influencing ribosome decoding and dynamics during translation elongation and are crucial for maintaining proteome integrity. In eukaryotes, wobble uridines are modified by Elongator, a large and highly conserved macromolecular complex. Elongator consists of two subcomplexes, namely Elp123 containing the enzymatically active Elp3 subunit and the associated Elp456 hetero-hexamer. The structure of the fully assembled complex and the function of the Elp456 subcomplex have remained elusive. Here, we show the cryo-electron microscopy structure of yeast Elongator at an overall resolution of 4.3 Å. We validate the obtained structure by complementary mutational analyses in vitro and in vivo. In addition, we determined various structures of the murine Elongator complex, including the fully assembled mouse Elongator complex at 5.9 Å resolution. Our results confirm the structural conservation of Elongator and its intermediates among eukaryotes. Furthermore, we complement our analyses with the biochemical characterization of the assembled human Elongator. Our results provide the molecular basis for the assembly of Elongator and its tRNA modification activity in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15623.map.gz emd_15623.map.gz | 18.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15623-v30.xml emd-15623-v30.xml emd-15623.xml emd-15623.xml | 26.2 KB 26.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15623_fsc.xml emd_15623_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_15623.png emd_15623.png | 83.8 KB | ||
| Filedesc metadata |  emd-15623.cif.gz emd-15623.cif.gz | 8.6 KB | ||
| Others |  emd_15623_half_map_1.map.gz emd_15623_half_map_1.map.gz emd_15623_half_map_2.map.gz emd_15623_half_map_2.map.gz | 225.5 MB 225.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15623 http://ftp.pdbj.org/pub/emdb/structures/EMD-15623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15623 | HTTPS FTP |
-Related structure data
| Related structure data |  8aswMC  8asvC  8at6C  8avgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15623.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15623.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
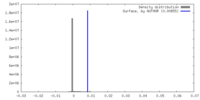
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15623_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
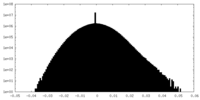
| Density Histograms |
-Half map: #2
| File | emd_15623_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
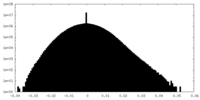
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast Elp123 in complex with alanine tRNA
| Entire | Name: Yeast Elp123 in complex with alanine tRNA |
|---|---|
| Components |
|
-Supramolecule #1: Yeast Elp123 in complex with alanine tRNA
| Supramolecule | Name: Yeast Elp123 in complex with alanine tRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 636.1 KDa |
-Macromolecule #1: Elongator complex protein 1
| Macromolecule | Name: Elongator complex protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 153.166266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVEHDKSGSK RQELRSNMRN LITLNKGKFK PTASTAEGDE DDLSFTLLDS VFDTLSDSIT CVLGSTDIGA IEVQQFMKDG SRNVLASFN IQTFDDKLLS FVHFADINQL VFVFEQGDII TATYDPVSLD PAETLIEIMG TIDNGIAAAQ WSYDEETLAM V TKDRNVVV ...String: MVEHDKSGSK RQELRSNMRN LITLNKGKFK PTASTAEGDE DDLSFTLLDS VFDTLSDSIT CVLGSTDIGA IEVQQFMKDG SRNVLASFN IQTFDDKLLS FVHFADINQL VFVFEQGDII TATYDPVSLD PAETLIEIMG TIDNGIAAAQ WSYDEETLAM V TKDRNVVV LSKLFEPISE YHLEVDDLKI SKHVTVGWGK KETQFRGKGA RAMEREALAS LKASGLVGNQ LRDPTMPYMV DT GDVTALD SHEITISWRG DCDYFAVSSV EEVPDEDDET KSIKRRAFRV FSREGQLDSA SEPVTGMEHQ LSWKPQGSLI ASI QRKTDL GEEDSVDVIF FERNGLRHGE FDTRLPLDEK VESVCWNSNS EALAVVLANR IQLWTSKNYH WYLKQELYAS DISY VKWHP EKDFTLMFSD AGFINIVDFA YKMAQGPTLE PFDNGTSLVV DGRTVNITPL ALANVPPPMY YRDFETPGNV LDVAC SFSN EIYAAINKDV LIFAAVPSIE EMKKGKHPSI VCEFPKSEFT SEVDSLRQVA FINDSIVGVL LDTDNLSRIA LLDIQD ITQ PTLITIVEVY DKIVLLRSDF DYNHLVYETR DGTVCQLDAE GQLMEITKFP QLVRDFRVKR VHNTSAEDDD NWSAESS EL VAFGITNNGK LFANQVLLAS AVTSLEITDS FLLFTTAQHN LQFVHLNSTD FKPLPLVEEG VEDERVRAIE RGSILVSV I PSKSSVVLQA TRGNLETIYP RIMVLAEVRK NIMAKRYKEA FIVCRTHRIN LDILHDYAPE LFIENLEVFI NQIGRVDYL NLFISCLSED DVTKTKYKET LYSGISKSFG MEPAPLTEMQ IYMKKKMFDP KTSKVNKICD AVLNVLLSNP EYKKKYLQTI ITAYASQNP QNLSAALKLI SELENSEEKD SCVTYLCFLQ DVNVVYKSAL SLYDVSLALL VAQKSQMDPR EYLPFLQELQ D NEPLRRKF LIDDYLGNYE KALEHLSEID KDGNVSEEVI DYVESHDLYK HGLALYRYDS EKQNVIYNIY AKHLSSNQMY TD AAVAYEM LGKLKEAMGA YQSAKRWREA MSIAVQKFPE EVESVAEELI SSLTFEHRYV DAADIQLEYL DNVKEAVALY CKA YRYDIA SLVAIKAKKD ELLEEVVDPG LGEGFGIIAE LLADCKGQIN SQLRRLRELR AKKEENPYAF YGQETEQADD VSVA PSETS TQESFFTRYT GKTGGTAKTG ASRRTAKNKR REERKRARGK KGTIYEEEYL VQSVGRLIER LNQTKPDAVR VVEGL CRRN MREQAHQIQK NFVEVLDLLK ANVKEIYSIS EKDRERVNEN GEVYYIPEIP VPEIHDFPKS HIVDF UniProtKB: Elongator complex protein 1 |
-Macromolecule #2: Elongator complex protein 2
| Macromolecule | Name: Elongator complex protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 89.51943 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVECITPEAI FIGANKQTQV SDIHKVKKIV AFGAGKTIAL WDPIEPNNKG VYATLKGHEA EVTCVRFVPD SDFMVSASED HHVKIWKFT DYSHLQCIQT IQHYSKTIVA LSALPSLISV GCADGTISIW RQNIQNDEFG LAHEFTIKKG FFYPLCLSLS K VEEKKYLL ...String: MVECITPEAI FIGANKQTQV SDIHKVKKIV AFGAGKTIAL WDPIEPNNKG VYATLKGHEA EVTCVRFVPD SDFMVSASED HHVKIWKFT DYSHLQCIQT IQHYSKTIVA LSALPSLISV GCADGTISIW RQNIQNDEFG LAHEFTIKKG FFYPLCLSLS K VEEKKYLL AIGGTNVNVF IASFILSDSG IEKCRVVAEL EGHEDWVKSL AFRHQETPGD YLLCSGSQDR YIRLWRIRIN DL IDDSEED SKKLTLLSNK QYKFQIDDEL RVGINFEALI MGHDDWISSL QWHESRLQLL AATADTSLMV WEPDETSGIW VCS LRLGEM SSKGASTATG SSGGFWSCLW FTHERMDFFL TNGKTGSWRM WATKDNIICD QRLGISGATK DVTDIAWSPS GEYL LATSL DQTTRLFAPW IYDASGRKRE IATWHEFSRP QIHGYDMICV ETVTDTRFVS GGDEKILRSF DLPKGVAGML QKFVG IQFE EKSEMPDSAT VPVLGLSNKA GEDDANEDDE EEEGGNKETP DITDPLSLLE CPPMEDQLQR HLLWPEVEKL YGHGFE ITC LDISPDQKLI ASACRSNNVQ NAVIRIFSTE NWLEIKPALP FHSLTITRLK FSKDGKFLLS VCRDRKWALW ERNMEDN TF ELRFKNEKPH TRIIWDADWA PLEFGNVFVT ASRDKTVKVW RHQKEPADDY VLEASIKHTK AVTAISIHDS MIREKILI S VGLENGEIYL YSYTLGKFEL ITQLNEDITP ADKITRLRWS HLKRNGKLFL GVGSSDLSTR IYSLAYE UniProtKB: Elongator complex protein 2 |
-Macromolecule #3: Elongator complex protein 3
| Macromolecule | Name: Elongator complex protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63.755059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARHGKGPKT NKKKLAPEKE RFIQCCADIT LELTDSLTSG TTREINLNGL ITKYSKKYKL KQQPRLTDII NSIPDQYKKY LLPKLKAKP VRTASGIAVV AVMCKPHRCP HIAYTGNICV YCPGGPDSDF EYSTQSYTGY EPTSMRAIRA RYDPYEQARG R VEQLKQLG ...String: MARHGKGPKT NKKKLAPEKE RFIQCCADIT LELTDSLTSG TTREINLNGL ITKYSKKYKL KQQPRLTDII NSIPDQYKKY LLPKLKAKP VRTASGIAVV AVMCKPHRCP HIAYTGNICV YCPGGPDSDF EYSTQSYTGY EPTSMRAIRA RYDPYEQARG R VEQLKQLG HSIDKVEYVL MGGTFMSLPK EYREDFIVKL HNALSGFNGN DIDEAILYSQ QSLTKCVGIT IETRPDYCTQ TH LDDMLKY GCTRLEIGVQ SLYEDVARDT NRGHTVRSVC ETFAVSKDAG YKVVSHMMPD LPNVGMERDI EQFKEYFENP DFR TDGLKI YPTLVIRGTG LYELWKTGRY KSYSANALVD LVARILALVP PWTRIYRVQR DIPMPLVTSG VDNGNLRELA LARM KDLGT TCRDVRTREV GIQEVHHKVQ PDQVELIRRD YYANGGWETF LSYEDPKKDI LIGLLRLRKA SKKYTYRKEF TSQRT SIVR ELHVYGSVVP LHSRDPRKFQ HQGFGTLLME EAERIAKEEH GSEKISVISG VGVRNYYGKL GYELDGPYMS KRI UniProtKB: Elongator complex protein 3 |
-Macromolecule #4: Alanine tRNA
| Macromolecule | Name: Alanine tRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.538932 KDa |
| Sequence | String: GGGCACAUGG CGCAGUUGGU AGCGCGCUUC CCUUGCAAGG AAGAGGUCAU CGGUUCGAUU CCGGUUGCGU CCA |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: 5'-DEOXYADENOSINE
| Macromolecule | Name: 5'-DEOXYADENOSINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: 5AD |
|---|---|
| Molecular weight | Theoretical: 251.242 Da |
| Chemical component information |  ChemComp-5AD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 15 s wait time, blot force 5, 5 s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 11080 / Average electron dose: 42.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | After initial rigid body fit, further fitting was done using MDFF and NAMDINATOR | ||||||
| Refinement | Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-8asw: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)