+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 305 A SynPspA rod after incubation with ATP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleotide binding / Helical assembly / ESCRT-III fold / Membrane remodeling / LIPID BINDING PROTEIN | |||||||||
| Function / homology | PspA/IM30 / PspA/IM30 family / Chloroplast membrane-associated 30 kD protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
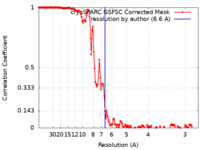
| Method | helical reconstruction / cryo EM / Resolution: 6.6 Å | |||||||||
 Authors Authors | Junglas B / Hudina E / Schoennenbeck P / Ritter I / Santiago-Schuebel B / Huesgen P / Sachse C | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies. Authors: Benedikt Junglas / Esther Hudina / Philipp Schönnenbeck / Ilona Ritter / Anja Heddier / Beatrix Santiago-Schübel / Pitter F Huesgen / Dirk Schneider / Carsten Sachse /  Abstract: Eukaryotic members of the endosome sorting complex required for transport-III (ESCRT-III) family have been shown to form diverse higher-order assemblies. The bacterial phage shock protein A (PspA) ...Eukaryotic members of the endosome sorting complex required for transport-III (ESCRT-III) family have been shown to form diverse higher-order assemblies. The bacterial phage shock protein A (PspA) has been identified as a member of the ESCRT-III superfamily, and PspA homo-oligomerizes to form rod-shaped assemblies. As observed for eukaryotic ESCRT-III, PspA forms tubular assemblies of varying diameters. Using electron cryo-electron microscopy, we determined 61 Synechocystis PspA structures and observed in molecular detail how the structural plasticity of PspA rods is mediated by conformational changes at three hinge regions in the monomer and by the fixed and changing molecular contacts between protomers. Moreover, we reduced and increased the structural plasticity of PspA rods by removing the loop connecting helices α3/α4 and the addition of nucleotides, respectively. Based on our analysis of PspA-mediated membrane remodeling, we suggest that the observed mode of structural plasticity is a prerequisite for the biological function of ESCRT-III members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15496.map.gz emd_15496.map.gz | 8.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15496-v30.xml emd-15496-v30.xml emd-15496.xml emd-15496.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15496_fsc.xml emd_15496_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_15496.png emd_15496.png | 49.4 KB | ||
| Masks |  emd_15496_msk_1.map emd_15496_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15496.cif.gz emd-15496.cif.gz | 5.4 KB | ||
| Others |  emd_15496_additional_1.map.gz emd_15496_additional_1.map.gz emd_15496_half_map_1.map.gz emd_15496_half_map_1.map.gz emd_15496_half_map_2.map.gz emd_15496_half_map_2.map.gz | 36.2 MB 226.1 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15496 http://ftp.pdbj.org/pub/emdb/structures/EMD-15496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15496 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15496 | HTTPS FTP |
-Validation report
| Summary document |  emd_15496_validation.pdf.gz emd_15496_validation.pdf.gz | 1005.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15496_full_validation.pdf.gz emd_15496_full_validation.pdf.gz | 1005.1 KB | Display | |
| Data in XML |  emd_15496_validation.xml.gz emd_15496_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  emd_15496_validation.cif.gz emd_15496_validation.cif.gz | 29 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15496 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15496 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15496 | HTTPS FTP |
-Related structure data
| Related structure data |  8akxMC  8akqC  8akrC  8aksC  8aktC  8akuC  8akvC  8akwC  8akyC  8akzC  8al0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15496.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15496.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.362 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15496_msk_1.map emd_15496_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local filtered map
| File | emd_15496_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15496_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15496_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Phage shock protein A (PspA)
| Entire | Name: Phage shock protein A (PspA) |
|---|---|
| Components |
|
-Supramolecule #1: Phage shock protein A (PspA)
| Supramolecule | Name: Phage shock protein A (PspA) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 162.2 kDa/nm |
-Macromolecule #1: Chloroplast membrane-associated 30 kD protein
| Macromolecule | Name: Chloroplast membrane-associated 30 kD protein / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.097758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSSAALEVLF QGPMELFNRV GRVLKSQLTH WQQQQEAPED LLERLLGEME LELIELRRAL AQTIATFKST ERQRDAQQL IAQRWYEKAQ AALDRGNEQL AREALGQRQS YQSHTEALGK SLGEQRALVE QVRGQLQKLE RKYLELKSQK N LYLARLKS ...String: MGSSHHHHHH SSSAALEVLF QGPMELFNRV GRVLKSQLTH WQQQQEAPED LLERLLGEME LELIELRRAL AQTIATFKST ERQRDAQQL IAQRWYEKAQ AALDRGNEQL AREALGQRQS YQSHTEALGK SLGEQRALVE QVRGQLQKLE RKYLELKSQK N LYLARLKS AIAAQKIEEI AGNLDNASAS SLFERIETKI LELEAERELL NPPPSPLDKK FEQWEEQQAV EATLAAMKAR RS LPPPSS UniProtKB: Chloroplast membrane-associated 30 kD protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)