[English] 日本語
 Yorodumi
Yorodumi- EMDB-15294: Cryo-EM structure of the strand transfer complex of the TnsB tran... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon) | |||||||||||||||
 Map data Map data | Map post-processed using DeepEMhancer and boxed out using Phenix. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Transposase / complex / CRISPR / transposition / DNA cleavage / ligation / DNA BINDING PROTEIN | |||||||||||||||
| Biological species |  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) | |||||||||||||||
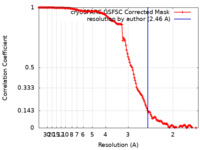
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | |||||||||||||||
 Authors Authors | Tenjo-Castano F / Sofos N / Lopez-Mendez B / Stutzke LS / Fuglsang A / Stella S / Montoya G | |||||||||||||||
| Funding support |  Denmark, 4 items Denmark, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon. Authors: Francisco Tenjo-Castaño / Nicholas Sofos / Blanca López-Méndez / Luisa S Stutzke / Anders Fuglsang / Stefano Stella / Guillermo Montoya /  Abstract: CRISPR-associated transposons (CASTs) are mobile genetic elements that co-opted CRISPR-Cas systems for RNA-guided transposition. Here we present the 2.4 Å cryo-EM structure of the Scytonema ...CRISPR-associated transposons (CASTs) are mobile genetic elements that co-opted CRISPR-Cas systems for RNA-guided transposition. Here we present the 2.4 Å cryo-EM structure of the Scytonema hofmannii (sh) TnsB transposase from Type V-K CAST, bound to the strand transfer DNA. The strand transfer complex displays an intertwined pseudo-symmetrical architecture. Two protomers involved in strand transfer display a catalytically competent active site composed by DDE residues, while other two, which play a key structural role, show active sites where the catalytic residues are not properly positioned for phosphodiester hydrolysis. Transposon end recognition is accomplished by the NTD1/2 helical domains. A singular in trans association of NTD1 domains of the catalytically competent subunits with the inactive DDE domains reinforces the assembly. Collectively, the structural features suggest that catalysis is coupled to protein-DNA assembly to secure proper DNA integration. DNA binding residue mutants reveal that lack of specificity decreases activity, but it could increase transposition in some cases. Our structure sheds light on the strand transfer reaction of DDE transposases and offers new insights into CAST transposition. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15294.map.gz emd_15294.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15294-v30.xml emd-15294-v30.xml emd-15294.xml emd-15294.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15294_fsc.xml emd_15294_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15294.png emd_15294.png | 88.5 KB | ||
| Masks |  emd_15294_msk_1.map emd_15294_msk_1.map | 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15294.cif.gz emd-15294.cif.gz | 7 KB | ||
| Others |  emd_15294_half_map_1.map.gz emd_15294_half_map_1.map.gz emd_15294_half_map_2.map.gz emd_15294_half_map_2.map.gz | 254.8 MB 254.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15294 http://ftp.pdbj.org/pub/emdb/structures/EMD-15294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15294 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15294.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15294.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map post-processed using DeepEMhancer and boxed out using Phenix. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15294_msk_1.map emd_15294_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15294_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15294_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of ShTnsB transposase with Strand Transfer Comple...
| Entire | Name: Ternary complex of ShTnsB transposase with Strand Transfer Complex DNA. |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of ShTnsB transposase with Strand Transfer Comple...
| Supramolecule | Name: Ternary complex of ShTnsB transposase with Strand Transfer Complex DNA. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 359 KDa |
-Macromolecule #1: TnsB
| Macromolecule | Name: TnsB / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 68.094609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNSQQNPDLA VHPLAIPMEG LLGESATTLE KNVIATQLSE EAQVKLEVIQ SLLEPCDRTT YGQKLREAAE KLNVSLRTVQ RLVKNWEQD GLVGLTQTSR ADKGKHRIGE FWENFITKTY KEGNKGSKRM TPKQVALRVE AKARELKDSK PPNYKTVLRV L APILEKQQ ...String: MNSQQNPDLA VHPLAIPMEG LLGESATTLE KNVIATQLSE EAQVKLEVIQ SLLEPCDRTT YGQKLREAAE KLNVSLRTVQ RLVKNWEQD GLVGLTQTSR ADKGKHRIGE FWENFITKTY KEGNKGSKRM TPKQVALRVE AKARELKDSK PPNYKTVLRV L APILEKQQ KAKSIRSPGW RGTTLSVKTR EGKDLSVDYS NHVWQCDHTR VDVLLVDQHG EILSRPWLTT VIDTYSRCIM GI NLGFDAP SSGVVALALR HAILPKRYGS EYKLHCEWGT YGKPEHFYTD GGKDFRSNHL SQIGAQLGFV CHLRDRPSEG GVV ERPFKT LNDQLFSTLP GYTGSNVQER PEDAEKDARL TLRELEQLLV RYIVDRYNQS IDARMGDQTR FERWEAGLPT VPVP IPERD LDICLMKQSR RTVQRGGCLQ FQNLMYRGEY LAGYAGETVN LRFDPRDITT ILVYRQENNQ EVFLTRAHAQ GLETE QLAL DEAEAASRRL RTAGKTISNQ SLLQEVVDRD ALVATKKSRK ERQKLEQTVL RSAAVDESNR ESLPSQIVEP DEVEST ETV HSQYEDIEVW DYEQLREEYG FGSEFELENL YFQ |
-Macromolecule #2: RE_Target
| Macromolecule | Name: RE_Target / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 24.402717 KDa |
| Sequence | String: (DA)(DT)(DA)(DA)(DG)(DG)(DA)(DT)(DT)(DT) (DT)(DA)(DC)(DT)(DG)(DA)(DT)(DG)(DA)(DC) (DA)(DA)(DT)(DA)(DA)(DT)(DT)(DT)(DG) (DT)(DC)(DA)(DC)(DA)(DA)(DC)(DG)(DA)(DC) (DA) (DT)(DA)(DT)(DA)(DA)(DT) ...String: (DA)(DT)(DA)(DA)(DG)(DG)(DA)(DT)(DT)(DT) (DT)(DA)(DC)(DT)(DG)(DA)(DT)(DG)(DA)(DC) (DA)(DA)(DT)(DA)(DA)(DT)(DT)(DT)(DG) (DT)(DC)(DA)(DC)(DA)(DA)(DC)(DG)(DA)(DC) (DA) (DT)(DA)(DT)(DA)(DA)(DT)(DT)(DA) (DG)(DT)(DC)(DA)(DC)(DT)(DG)(DT)(DA)(DC) (DA)(DC) (DG)(DT)(DA)(DG)(DA)(DG)(DA) (DC)(DG)(DT)(DA)(DG)(DC)(DA)(DA)(DT)(DG) (DC)(DT) |
-Macromolecule #3: RE_PolyA
| Macromolecule | Name: RE_PolyA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 22.876809 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DT)(DG)(DT)(DA)(DC) (DA)(DG)(DT)(DG)(DA)(DC)(DT)(DA)(DA) (DT)(DT)(DA)(DT)(DA)(DT)(DG)(DT)(DC)(DG) (DT) (DT)(DG)(DT)(DG)(DA)(DC) ...String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DT)(DG)(DT)(DA)(DC) (DA)(DG)(DT)(DG)(DA)(DC)(DT)(DA)(DA) (DT)(DT)(DA)(DT)(DA)(DT)(DG)(DT)(DC)(DG) (DT) (DT)(DG)(DT)(DG)(DA)(DC)(DA)(DA) (DA)(DT)(DT)(DA)(DT)(DT)(DG)(DT)(DC)(DA) (DT)(DC) (DA)(DG)(DT)(DA)(DA)(DA)(DA) (DT)(DC)(DC)(DT)(DT)(DA)(DT) |
-Macromolecule #4: Target_1
| Macromolecule | Name: Target_1 / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 4.559971 KDa |
| Sequence | String: (DA)(DG)(DC)(DA)(DT)(DT)(DG)(DC)(DT)(DA) (DC)(DG)(DT)(DC)(DT) |
-Macromolecule #5: LE_Target
| Macromolecule | Name: LE_Target / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 24.63483 KDa |
| Sequence | String: (DA)(DA)(DT)(DT)(DA)(DA)(DA)(DT)(DA)(DG) (DT)(DC)(DA)(DC)(DA)(DA)(DT)(DG)(DA)(DC) (DA)(DT)(DT)(DA)(DA)(DT)(DC)(DT)(DG) (DT)(DC)(DA)(DC)(DC)(DG)(DA)(DC)(DG)(DA) (DC) (DA)(DG)(DA)(DT)(DA)(DA) ...String: (DA)(DA)(DT)(DT)(DA)(DA)(DA)(DT)(DA)(DG) (DT)(DC)(DA)(DC)(DA)(DA)(DT)(DG)(DA)(DC) (DA)(DT)(DT)(DA)(DA)(DT)(DC)(DT)(DG) (DT)(DC)(DA)(DC)(DC)(DG)(DA)(DC)(DG)(DA) (DC) (DA)(DG)(DA)(DT)(DA)(DA)(DT)(DT) (DT)(DG)(DT)(DC)(DA)(DC)(DT)(DG)(DT)(DA) (DC)(DA) (DC)(DT)(DA)(DC)(DG)(DC)(DC) (DT)(DT)(DT)(DT)(DG)(DT)(DG)(DG)(DA)(DG) (DA)(DT)(DG) |
-Macromolecule #6: LE_PolyA
| Macromolecule | Name: LE_PolyA / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 23.238008 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DT)(DG)(DT)(DA)(DC) (DA)(DG)(DT)(DG)(DA)(DC)(DA)(DA)(DA) (DT)(DT)(DA)(DT)(DC)(DT)(DG)(DT)(DC)(DG) (DT) (DC)(DG)(DG)(DT)(DG)(DA) ...String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DT)(DG)(DT)(DA)(DC) (DA)(DG)(DT)(DG)(DA)(DC)(DA)(DA)(DA) (DT)(DT)(DA)(DT)(DC)(DT)(DG)(DT)(DC)(DG) (DT) (DC)(DG)(DG)(DT)(DG)(DA)(DC)(DA) (DG)(DA)(DT)(DT)(DA)(DA)(DT)(DG)(DT)(DC) (DA)(DT) (DT)(DG)(DT)(DG)(DA)(DC)(DT) (DA)(DT)(DT)(DT)(DA)(DA)(DT)(DT) |
-Macromolecule #7: Target_2
| Macromolecule | Name: Target_2 / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 4.54699 KDa |
| Sequence | String: (DC)(DA)(DT)(DC)(DT)(DC)(DC)(DA)(DC)(DA) (DA)(DA)(DA)(DG)(DG) |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4728 / Average exposure time: 40.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8aa5: |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)