[English] 日本語
 Yorodumi
Yorodumi- EMDB-13977: Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated) | ||||||||||||||||||
 Map data Map data | phenix density modified map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | supercomplex / respiratory chain / cytochrome bcc-aa3 supercomplex / proton translocation / bioenergetics / membrane protein / electron transport / OXIDOREDUCTASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationthiamine biosynthetic process / aerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport ...thiamine biosynthetic process / aerobic electron transport chain / cytochrome-c oxidase / oxidative phosphorylation / quinol-cytochrome-c reductase / quinol-cytochrome-c reductase activity / cytochrome-c oxidase activity / electron transport coupled proton transport / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / ATP synthesis coupled electron transport / respiratory electron transport chain / monooxygenase activity / electron transport chain / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Corynebacterium glutamicum ATCC 13032 (bacteria) Corynebacterium glutamicum ATCC 13032 (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||||||||
 Authors Authors | Kao W-C / Hunte C | ||||||||||||||||||
| Funding support |  Germany, Germany,  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for safe and efficient energy conversion in a respiratory supercomplex. Authors: Wei-Chun Kao / Claire Ortmann de Percin Northumberland / Tat Cheung Cheng / Julio Ortiz / Alexandre Durand / Ottilie von Loeffelholz / Oliver Schilling / Martin L Biniossek / Bruno P Klaholz / Carola Hunte /   Abstract: Proton-translocating respiratory complexes assemble into supercomplexes that are proposed to increase the efficiency of energy conversion and limit the production of harmful reactive oxygen species ...Proton-translocating respiratory complexes assemble into supercomplexes that are proposed to increase the efficiency of energy conversion and limit the production of harmful reactive oxygen species during aerobic cellular respiration. Cytochrome bc complexes and cytochrome aa oxidases are major drivers of the proton motive force that fuels ATP generation via respiration, but how wasteful electron- and proton transfer is controlled to enhance safety and efficiency in the context of supercomplexes is not known. Here, we address this question with the 2.8 Å resolution cryo-EM structure of the cytochrome bcc-aa (III-IV) supercomplex from the actinobacterium Corynebacterium glutamicum. Menaquinone, substrate mimics, lycopene, an unexpected Q site, dioxygen, proton transfer routes, and conformational states of key protonable residues are resolved. Our results show how safe and efficient energy conversion is achieved in a respiratory supercomplex through controlled electron and proton transfer. The structure may guide the rational design of drugs against actinobacteria that cause diphtheria and tuberculosis. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13977.map.gz emd_13977.map.gz | 26.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13977-v30.xml emd-13977-v30.xml emd-13977.xml emd-13977.xml | 38.3 KB 38.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13977.png emd_13977.png | 146.2 KB | ||
| Filedesc metadata |  emd-13977.cif.gz emd-13977.cif.gz | 10 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13977 http://ftp.pdbj.org/pub/emdb/structures/EMD-13977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13977 | HTTPS FTP |
-Related structure data
| Related structure data |  7qhoMC  7qhmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13977.map.gz / Format: CCP4 / Size: 28.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13977.map.gz / Format: CCP4 / Size: 28.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phenix density modified map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
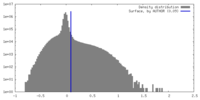
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : cytochrome bcc-aa3 supercomplex
+Supramolecule #1: cytochrome bcc-aa3 supercomplex
+Macromolecule #1: Cytochrome bc1 complex Rieske iron-sulfur subunit
+Macromolecule #2: Cytochrome bc1 complex cytochrome b subunit
+Macromolecule #3: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #4: Cytochrome c oxidase subunit 1
+Macromolecule #5: Cytochrome c oxidase subunit 2
+Macromolecule #6: Cytochrome c oxidase subunit 3
+Macromolecule #7: Cytochrome c oxidase polypeptide 4
+Macromolecule #8: Uncharacterized protein Cgl2664/cg2949
+Macromolecule #9: Uncharacterized membrane protein Cgl2017/cg2211
+Macromolecule #10: Hypothetical membrane protein
+Macromolecule #11: Actinobacterial supercomplex, subunit C (AscC)
+Macromolecule #12: Hypothetical membrane protein
+Macromolecule #13: Thiamine biosynthesis protein X
+Macromolecule #14: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #15: [(2~{R})-3-[[(1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-[(2~{R},3~{S...
+Macromolecule #16: (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3...
+Macromolecule #17: MENAQUINONE-9
+Macromolecule #18: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #19: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #20: CARDIOLIPIN
+Macromolecule #21: LYCOPENE
+Macromolecule #22: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #23: HEME C
+Macromolecule #24: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
+Macromolecule #25: HEME-AS
+Macromolecule #26: COPPER (II) ION
+Macromolecule #27: CALCIUM ION
+Macromolecule #28: MANGANESE (II) ION
+Macromolecule #29: DINUCLEAR COPPER ION
+Macromolecule #30: DIACYL GLYCEROL
+Macromolecule #31: PALMITIC ACID
+Macromolecule #32: [(2~{R})-3-[[(1~{S},2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S...
+Macromolecule #33: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3453 / Average electron dose: 49.95 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7qho: |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)