+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S.c. Condensin core in DNA- and ATP-bound state | |||||||||
 Map data Map data | deepEMhancer processed cryoEM map S.c. Condensin in DNA- and ATP-bound state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SMC-motor protein / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of meiotic DNA double-strand break formation / meiotic chromosome condensation / Condensation of Prometaphase Chromosomes / tRNA gene clustering / meiotic chromosome separation / condensin complex / DNA secondary structure binding / maintenance of rDNA / rDNA chromatin condensation / nucleophagy ...negative regulation of meiotic DNA double-strand break formation / meiotic chromosome condensation / Condensation of Prometaphase Chromosomes / tRNA gene clustering / meiotic chromosome separation / condensin complex / DNA secondary structure binding / maintenance of rDNA / rDNA chromatin condensation / nucleophagy / synaptonemal complex assembly / condensed chromosome, centromeric region / mitotic chromosome condensation / chromosome condensation / silent mating-type cassette heterochromatin formation / minor groove of adenine-thymine-rich DNA binding / mitotic sister chromatid segregation / condensed chromosome / histone binding / double-stranded DNA binding / cell division / chromatin binding / chromatin / nucleolus / ATP hydrolysis activity / mitochondrion / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
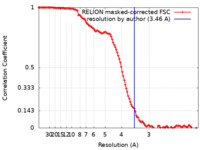
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Lecomte L / Hassler M / Haering C / Eustermann S | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: A hold-and-feed mechanism drives directional DNA loop extrusion by condensin. Authors: Indra A Shaltiel / Sumanjit Datta / Léa Lecomte / Markus Hassler / Marc Kschonsak / Sol Bravo / Catherine Stober / Jenny Ormanns / Sebastian Eustermann / Christian H Haering /  Abstract: Structural maintenance of chromosomes (SMC) protein complexes structure genomes by extruding DNA loops, but the molecular mechanism that underlies their activity has remained unknown. We show that ...Structural maintenance of chromosomes (SMC) protein complexes structure genomes by extruding DNA loops, but the molecular mechanism that underlies their activity has remained unknown. We show that the active condensin complex entraps the bases of a DNA loop transiently in two separate chambers. Single-molecule imaging and cryo-electron microscopy suggest a putative power-stroke movement at the first chamber that feeds DNA into the SMC-kleisin ring upon adenosine triphosphate binding, whereas the second chamber holds on upstream of the same DNA double helix. Unlocking the strict separation of "motor" and "anchor" chambers turns condensin from a one-sided into a bidirectional DNA loop extruder. We conclude that the orientation of two topologically bound DNA segments during the SMC reaction cycle determines the directionality of DNA loop extrusion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13934.map.gz emd_13934.map.gz | 106.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13934-v30.xml emd-13934-v30.xml emd-13934.xml emd-13934.xml | 35.5 KB 35.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13934_fsc.xml emd_13934_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_13934.png emd_13934.png | 72.1 KB | ||
| Masks |  emd_13934_msk_1.map emd_13934_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13934.cif.gz emd-13934.cif.gz | 10.4 KB | ||
| Others |  emd_13934_additional_1.map.gz emd_13934_additional_1.map.gz emd_13934_additional_2.map.gz emd_13934_additional_2.map.gz emd_13934_half_map_1.map.gz emd_13934_half_map_1.map.gz emd_13934_half_map_2.map.gz emd_13934_half_map_2.map.gz | 98.6 MB 76.4 MB 98.6 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13934 http://ftp.pdbj.org/pub/emdb/structures/EMD-13934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13934 | HTTPS FTP |
-Validation report
| Summary document |  emd_13934_validation.pdf.gz emd_13934_validation.pdf.gz | 832.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13934_full_validation.pdf.gz emd_13934_full_validation.pdf.gz | 832 KB | Display | |
| Data in XML |  emd_13934_validation.xml.gz emd_13934_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_13934_validation.cif.gz emd_13934_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13934 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13934 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13934 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13934 | HTTPS FTP |
-Related structure data
| Related structure data |  7qenMC  7qfwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13934.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13934.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer processed cryoEM map S.c. Condensin in DNA- and ATP-bound state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
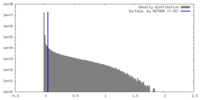
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13934_msk_1.map emd_13934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
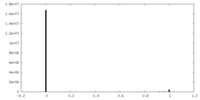
| Density Histograms |
-Additional map: unsharpened cryoEM map S.c. Condensin in DNA- and ATP-bound state
| File | emd_13934_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened cryoEM map S.c. Condensin in DNA- and ATP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
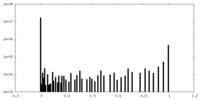
| Density Histograms |
-Additional map: local resolution filtered and sharpened cryoEM map S.c....
| File | emd_13934_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered and sharpened cryoEM map S.c. Condensin in DNA- and ATP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryoEM half map 1 S.c. Condensin in DNA- and ATP-bound state
| File | emd_13934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM half map 1 S.c. Condensin in DNA- and ATP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
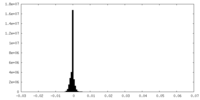
| Density Histograms |
-Half map: cryoEM half map 1 S.c. Condensin in DNA- and ATP-bound state
| File | emd_13934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM half map 1 S.c. Condensin in DNA- and ATP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP- and DNA- bound Sc Condensin core complex
| Entire | Name: ATP- and DNA- bound Sc Condensin core complex |
|---|---|
| Components |
|
-Supramolecule #1: ATP- and DNA- bound Sc Condensin core complex
| Supramolecule | Name: ATP- and DNA- bound Sc Condensin core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 645 kDa/nm |
-Macromolecule #1: DNA (35-MER)
| Macromolecule | Name: DNA (35-MER) / type: dna / ID: 1 Details: 50-mer DNA ligand (sequence below) was modelled as poly-dA due to missing sequence register 5'-GTTGACAGTG TCGCAACCTG CACAGGCAAG CTGCTGAGTC TGGTGTAGAC-3' Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.615376 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA) |
-Macromolecule #2: DNA (35-MER)
| Macromolecule | Name: DNA (35-MER) / type: dna / ID: 2 Details: 50-mer DNA ligand (sequence below) was modelled as poly-dT due to missing sequence register. 5'-GTCTACACCAGACTCAGCAGCTTGCCTGTGCAGGTTGCGACACTGTCAAC-3' Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.164683 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) |
-Macromolecule #3: Structural maintenance of chromosomes protein 4
| Macromolecule | Name: Structural maintenance of chromosomes protein 4 / type: protein_or_peptide / ID: 3 Details: ATPase deficient Walker B motif mutant central coiled-coil region results in faulty sequence alignment as it was not build in the structure To circumvent this technical problem, we had to ...Details: ATPase deficient Walker B motif mutant central coiled-coil region results in faulty sequence alignment as it was not build in the structure To circumvent this technical problem, we had to delete the sequence corresponding to the coiled coil regions. This error needs to be corrected in communication with the pdb curator.,ATPase deficient Walker B motif mutant central coiled-coil region results in faulty sequence alignment as it was not build in the structure To circumvent this technical problem, we had to delete the sequence corresponding to the coiled coil regions. This error needs to be corrected in communication with the pdb curator.,ATPase deficient Walker B motif mutant central coiled-coil region results in faulty sequence alignment as it was not build in the structure To circumvent this technical problem, we had to delete the sequence corresponding to the coiled coil regions. This error needs to be corrected in communication with the pdb curator. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 167.716125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDSPLSKRQ KRKAAQEPEL SLDQGDAEEE SQVENRVNLS ENTPEPDLPA LEASYSKSYT PRKLVLSSGE NRYAFSQPTN STTTSLHVP NLQPPKTSSR GRDHKSYSQS PPRSPGRSPT RRLEL(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK) ...String: MSDSPLSKRQ KRKAAQEPEL SLDQGDAEEE SQVENRVNLS ENTPEPDLPA LEASYSKSYT PRKLVLSSGE NRYAFSQPTN STTTSLHVP NLQPPKTSSR GRDHKSYSQS PPRSPGRSPT RRLEL(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)YDSHQSSS KQQSRLFINE LVLENFKSYA GKQVVGP FH TSFSAVVGPN GSGKSNVIDS MLFVFGFRAN KMRQDRLSDL IHKSEAFPSL QSCSVAVHFQ YVIDESSGTS RIDEEKPG L IITRKAFKNN SSKYYINEKE SSYTEVTKLL KNEGIDLDHK RFLILQGEVE NIAQMKPKAE KESDDGLLEY LEDIIGTAN YKPLIEERMG QIENLNEVCL EKENRFEIVD REKNSLESGK ETALEFLEKE KQLTLLRSKL FQFKLLQSNS KLASTLEKIS SSNKDLEDE RMKFQESLKK VDEIKAQRKE IKDRISSCSS KEKTLVLERR ELEGTRVSLE ERTKNLVSKM EKAEKTLKST K HSISEAEN MLEELRGQQT EHETEIKDLT QLLEKERSIL DDIKLSLKDK TKDISAEIIR HEKELEPWDL QLQEKESQIQ LA ESELSLL EETQAKLKKN VETLEEKILA KKTHKQELQD LILDLKKKLN SLKDERSQGE KNFTSAHLKL KEMQKVLNAH RQR AMEARS SLSKAQNKSK VLTALSRLQK SGRINGFHGR LGDLGAIDDS FDIAISTACP RLDDVVVDTV ECAQHCIDYL RKNK LGYAR FILLDRLRQF NLQPISTPEN VPRLFDLVKP KNPKFSNAFY SVLRDTLVAQ NLKQANNVAY GKKRFRVVTV DGKLI DISG TMSGGGNHVA KGLMKLGTNQ SDKVDDYTPE EVDKIERELS ERENNFRVAS DTVHEMEEEL KKLRDHEPDL ESQISK AEM EADSLASELT LAEQQVKEAE MAYVKAVSDK AQLNVVMKNL ERLRGEYNDL QSETKTKKEK IKGLQDEIMK IGGIKLQ MQ NSKVESVCQK LDILVAKLKK VKSASKKSGG DVVKFQKLLQ NSERDVELSS NELKVIEEQL KHTKLALAEN DTNMTETL N LKVELKEQSE QLKEQMEDME ESINEFKSIE IEMKNKLEKL NSLLTYIKSE ITQQEKGLNE LSIRDVTHTL GMLDDNKMD SVKEDVKNNQ ELDQEYRSCE TQDESEIKDD ETSCDNYHPM NVDETSDEVS RGIPRLSEDE LRELDVELIE SKINELSYYV EETNVDIGV LEEYARRLAE FKRRKLDLNN AVQKRDEVKE QLGILKKKRF DEFMAGFNII SMTLKEMYQM ITMGGNAELE L VDSLDPFS EGVTFSVMPP KKSWRNITNL SGGEKTLSSL ALVFALHKYK PTPLYVMDQI DAALDFRNVS IVANYIKERT KN AQFIVIS LRNNMFELAQ QLVGVYKRDN RTKSTTIKNI DILNRTRIPG LINGATGWSH PQFEKAGGGS GGGSGGGSWS HPQ FEKGGG SGGGSGGGSW SHPQFEK UniProtKB: Structural maintenance of chromosomes protein 4, Structural maintenance of chromosomes protein 4 |
-Macromolecule #4: Structural maintenance of chromosomes protein 2
| Macromolecule | Name: Structural maintenance of chromosomes protein 2 / type: protein_or_peptide / ID: 4 / Details: ATPase deficient Walker B motif mutant / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.124891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVEELIIDG FKSYATRTVI TDWDPQFNAI TGLNGSGKSN ILDAICFVLG IASMSTVRAS SLQDLIYKRG QAGVTKASVT IVFDNTDKS NSPIGFTNSP QISVTRQVVL GGTSKYLING HRAPQQSVLQ LFQSVQLNIN NPNFLIMQGK ITKVLNMKPS E ILSLIEEA ...String: MKVEELIIDG FKSYATRTVI TDWDPQFNAI TGLNGSGKSN ILDAICFVLG IASMSTVRAS SLQDLIYKRG QAGVTKASVT IVFDNTDKS NSPIGFTNSP QISVTRQVVL GGTSKYLING HRAPQQSVLQ LFQSVQLNIN NPNFLIMQGK ITKVLNMKPS E ILSLIEEA AGTKMFEDRR EKAERTMSKK ETKLQENRTL LTEEIEPKLE KLRNEKRMFL EFQSTQTDLE KTERIVVSYE YY NIKHKHT SIRETLENGE TRMKMLNEFV KKTSEEIDSL NEDVEEIKLQ KEKELHKEGT ISKLENKENG LLNEISRLKT SLS IKVENL NDTTEKSKAL ESEIASSSAK LIEKKSAYAN TEKDYKMVQE QLSKQRDLYK RKEELVSTLT TGISSTGAAD GGYN AQLAK AKTELNEVSL AIKKSSMKME LLKKELLTIE PKLKEATKDN ELNVKHVKQC QETCDKLRAR LVEYGFDPSR IKDLK QRED KLKSHYYQTC KNSEYLKRRV TNLEFNYTKP YPNFEASFVH GVVGQLFQID NDNIRYATAL QTCAGGRLFN VVVQDS QTA TQLLERGRLR KRVTIIPLDK IYTRPISSQV LDLAKKIAPG KVELAINLIR FDESITKAME FIFGNSLICE DPETAKK IT FHPKIRARSI TLQGDVYDPE GTLSGGSRNT SESLLVDIQK YNQIQKQIET IQADLNHVTE ELQTQYATSQ KTKTIQSD L NLSLHKLDLA KRNLDANPSS QIIARNEEIL RDIGECENEI KTKQMSLKKC QEEVSTIEKD MKEYDSDKGS KLNELKKEL KLLAKELEEQ ESESERKYDL FQNLELETEQ LSSELDSNKT LLHNHLKSIE SLKLENSDLE GKIRGVEDDL VTVQTELNEE KKRLMDIDD ELNELETLIK KKQDEKKSSE LELQKLVHDL NKYKSNTNNM EKIIEDLRQK HEFLEDFDLV RNIVKQNEGI D LDTYRERS KQLNEKFQEL RKKVNPNIMN MIENVEKKEA ALKTMIKTIE KDKMKIQETI SKLNEYKRET LVKTWEKVTL DF GNIFADL LPNSFAKLVP CEGKDVTQGL EVKVKLGNIW KESLIELSGG QRSLIALSLI MALLQFRPAP MYILDQVDAA LDL SHTQNI GHLIKTRFKG SQFIVVSLKE GMFANANRVF RTRFQDGTSV VSIM UniProtKB: Structural maintenance of chromosomes protein 2 |
-Macromolecule #5: Condensin complex subunit 2
| Macromolecule | Name: Condensin complex subunit 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.721219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV ...String: MTTQLRYENN DDDERVEYNL FTNRSTMMAN FEEWIKMATD NKINSRNSWN FALIDYFYDL DVLKDGENNI NFQKASATLD GCIKIYSSR VDSVTTETGK LLSGLAQRKT NGASNGDDSN GGNGEGLGGD SDEANIEIDP LTGMPISNDP DVNNTRRRVY N RVLETTLV EFETIKMKEL DQELIIDPLF KKALVDFDEG GAKSLLLNTL NIDNTARVIF DASIKDTQNV GQGKLQRKEE EL IERDSLV DDENEPSQSL ISTRNDSTVN DSVISAPSME DEILSLGMDF IKFDQIAVCE ISGSIEQLRN VVEDINQAKD FIE NVNNRF DNFLTEEELQ AAVPDNAEDD SDGFDMGMQQ ELCYPDENHD NTSHDEQDDD NVNSTTGSIF EKDLMAYFDE NLNR NWRGR EHWKVRNFKK ANLVNKESDL LEETRTTIGD TTDKNTTDDK SMDTKKKHKQ KKVLEIDFFK TDDSFEDKVF ASKGR TKID MPIKNRKNDT HYLLPDDFHF STDRITRLFI KPAQKMSLFS HRKHTRGDVS SGLFEKSTVS ANHSNNDIPT IADEHF WAD NYERKEQEEK EKEQSKEVGD VVGGALDNPF EDDMDGVDFN QAFEGTDDNE EASVKLDLQD DEDHKFPIRE NKVTYSR VS KKVDVRRLKK NVWRSINNLI QEHDSRKNRE QSSNDSETHT EDESTKELKF SDIIQGISKM YSDDTLKDIS TSFCFICL L HLANEHGLQI THTENYNDLI VNYEDLATTQ AASLVGGGHH RPHHGGHHHH HHGGRIFYPY DVPDYAGYPY DVPDYAGSY PYDVPNYAAG H UniProtKB: Condensin complex subunit 2 |
-Macromolecule #6: Condensin complex subunit 1
| Macromolecule | Name: Condensin complex subunit 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.116766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGFSLSEYL TKFQTTDRES YPRLQDPSRE LNVIIDQLAV SPEQIDASPD SLEALIDLCH DFPHLTPKLQ TQLSYLISSS LSNLSKDIK ANLSSNVNFT EIGGLIPQWK RHLEEYGYLI QVLLTFLQDE LHKVSSQSTN LNRSAKNSKN DSANVELFKR D CNQMENLL ...String: MSGFSLSEYL TKFQTTDRES YPRLQDPSRE LNVIIDQLAV SPEQIDASPD SLEALIDLCH DFPHLTPKLQ TQLSYLISSS LSNLSKDIK ANLSSNVNFT EIGGLIPQWK RHLEEYGYLI QVLLTFLQDE LHKVSSQSTN LNRSAKNSKN DSANVELFKR D CNQMENLL ESITKLLEIN LSKIFQTTPE KDLFIGLFTR PLFVLLEIEP VTKVSSLKMF IQRILAMCVK NHGQSSSIQS SL MTNLTYF LHLSVFNAEL LKLLNDEYNY PQLTEDILKE ISTRVFNAKD TTGPKAISNF LIKLSELSPG IMLRQMNLVI TLL NNSSIT LRCSVVEACG NIVAELAQDP QTMEHYKQQI AVLIELLEER FQDSNPYVRT KAIQGCSKIC DLSSKFNKSK AKFT SLAVR SLQDRSSLVR RNSVKLLSKL LLKHPFKAIH GSQLRLSEWE EYLKGSESQL NSTLKKVESQ ETLNDTIERS LIEEE VEQD EGQCRTELEG SFNKSAELSR IENEVENINA TNTSVLMKLK LMIVYYKDAI SFIKEIHKSI ELISNLLFSK NRNEVL ESM DFLVLADAFD IELSEFGIKK MLHLVWMKGT NDEGTSISVH LIECYKQLFL TAPDSCNMQE KAAHIAKNLI NLSIGAS IA DLASLEQLLG MMYEQKLIDQ HVINILWAIY NSASKASMQK EQNVNNRDSE KGFSKEQIHG SIIILGMLSL ADNEIALK G LESLLNIGLG AVGLKDLTLC RYSCLALERM VPKRSTIITK AINQELEDVA VKKLYAIIIN YTKDNEYYPM CEQALSALF TISSKPDILA TDLIREKTMM TFGKPEEEDS ILSLEQSSRV VSLSQLLFIV GQVAIKTLVY LEKCEAEFKK RKIEAETRNG KVKNQGADV TNTTQDNGGD KELEMIGGTN EDDFTDAIQF VKENELLFGE KSILGKFCPI VEEIVSNSSR FSDPMLQRTA T LCLEKLMC LSSKYCEKSL PLLITVMEKS PDPTIRSNAV LGLGDMAVCF NNLVDENTDY LYRRLHDENL MVQRTCLMTV TF LILAGQV KVKGQLGEMA KCLDNPDQGI SDMCRLFFTE LASKDNAIYN GFIDIFSNLS SDDLLGKESF KKIIKFLLTF IDK ERHQKQ LNEKLVGRLR KCETQKQWDD IAFVLNNLPY KNEDVTALLE QGFKVVSAKE UniProtKB: Condensin complex subunit 1 |
-Macromolecule #7: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.645 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 2 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 6544 / Average exposure time: 8.0 sec. / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)