[English] 日本語
 Yorodumi
Yorodumi- EMDB-13895: CryoEM structure of the Smc5/6-holocomplex (composite structure) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the Smc5/6-holocomplex (composite structure) | |||||||||
 Map data Map data | CryoEM structure of the Smc5/6 holocomplex; composite map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Structural Maintenance of Chromosomes / SMC / holo-complex / recombination / stalled replication fork | |||||||||
| Function / homology |  Function and homology information Function and homology informationSmc5-Smc6 complex / resolution of DNA recombination intermediates / SUMO ligase activity / chromosome separation / DNA double-strand break attachment to nuclear envelope / Platelet degranulation / SUMOylation of DNA damage response and repair proteins / chromatin looping / Transferases; Acyltransferases; Aminoacyltransferases / SUMO transferase activity ...Smc5-Smc6 complex / resolution of DNA recombination intermediates / SUMO ligase activity / chromosome separation / DNA double-strand break attachment to nuclear envelope / Platelet degranulation / SUMOylation of DNA damage response and repair proteins / chromatin looping / Transferases; Acyltransferases; Aminoacyltransferases / SUMO transferase activity / DNA damage tolerance / recombinational repair / regulation of telomere maintenance / protein sumoylation / protein serine/threonine kinase inhibitor activity / double-strand break repair via homologous recombination / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / nuclear envelope / single-stranded DNA binding / site of double-strand break / damaged DNA binding / chromosome, telomeric region / DNA repair / ATP hydrolysis activity / mitochondrion / zinc ion binding / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.0 Å | |||||||||
 Authors Authors | Hallett ST / Oliver AW | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Cryo-EM structure of the Smc5/6 holo-complex. Authors: Stephen T Hallett / Isabella Campbell Harry / Pascale Schellenberger / Lihong Zhou / Nora B Cronin / Jonathan Baxter / Thomas J Etheridge / Johanne M Murray / Antony W Oliver /  Abstract: The Smc5/6 complex plays an essential role in the resolution of recombination intermediates formed during mitosis or meiosis, or as a result of the cellular response to replication stress. It also ...The Smc5/6 complex plays an essential role in the resolution of recombination intermediates formed during mitosis or meiosis, or as a result of the cellular response to replication stress. It also functions as a restriction factor preventing viral replication. Here, we report the cryogenic EM (cryo-EM) structure of the six-subunit budding yeast Smc5/6 holo-complex, reconstituted from recombinant proteins expressed in insect cells - providing both an architectural overview of the entire complex and an understanding of how the Nse1/3/4 subcomplex binds to the hetero-dimeric SMC protein core. In addition, we demonstrate that a region within the head domain of Smc5, equivalent to the 'W-loop' of Smc4 or 'F-loop' of Smc1, mediates an important interaction with Nse1. Notably, mutations that alter the surface-charge profile of the region of Nse1 which accepts the Smc5-loop, lead to a slow-growth phenotype and a global reduction in the chromatin-associated fraction of the Smc5/6 complex, as judged by single molecule localisation microscopy experiments in live yeast. Moreover, when taken together, our data indicates functional equivalence between the structurally unrelated KITE and HAWK accessory subunits associated with SMC complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13895.map.gz emd_13895.map.gz | 106.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13895-v30.xml emd-13895-v30.xml emd-13895.xml emd-13895.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13895.png emd_13895.png | 30 KB | ||
| Filedesc metadata |  emd-13895.cif.gz emd-13895.cif.gz | 8.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13895 http://ftp.pdbj.org/pub/emdb/structures/EMD-13895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13895 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13895 | HTTPS FTP |
-Related structure data
| Related structure data |  7qcdMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13895.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13895.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of the Smc5/6 holocomplex; composite map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
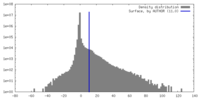
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Smc5/6 holo-complex
| Entire | Name: Smc5/6 holo-complex |
|---|---|
| Components |
|
-Supramolecule #1: Smc5/6 holo-complex
| Supramolecule | Name: Smc5/6 holo-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Structural maintenance of chromosomes protein 5
| Macromolecule | Name: Structural maintenance of chromosomes protein 5 / type: protein_or_peptide / ID: 1 Details: Saccharomyces cerevisiase Smc5 fused to C-terminal FLAG-tag. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 126.23618 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ ...String: MTSLIDLGRY VERTHHGEDT EPRSKRVKIA KPDLSSFQPG SIIKIRLQDF VTYTLTEFNL SPSLNMIIGP NGSGKSTFVC AVCLGLAGK PEYIGRSKKV EDFIKNGQDV SKIEITLKNS PNVTDIEYID ARDETIKITR IITRSKRRSD YLINDYQVSE S VVKTLVAQ LNIQLDNLCQ FLSQERVEEF ARLKSVKLLV ETIRSIDASL LDVLDELREL QGNEQSLQKD LDFKKAKIVH LR QESDKLR KSVESLRDFQ NKKGEIELHS QLLPYVKVKD HKEKLNIYKE EYERAKANLR AILKDKKPFA NTKKTLENQV EEL TEKCSL KTDEFLKAKE KINEIFEKLN TIRDEVIKKK NQNEYYRGRT KKLQATIIST KEDFLRSQEI LAQTHLPEKS VFED IDIKR KEIINKEGEI RDLISEIDAK ANAINHEMRS IQRQAESKTK SLTTTDKIGI LNQDQDLKEV RDAVLMVREH PEMKD KILE PPIMTVSAIN AQFAAYLAQC VDYNTSKALT VVDSDSYKLF ANPILDKFKV NLRELSSADT TPPVPAETVR DLGFEG YLS DFITGDKRVM KMLCQTSKIH TIPVSRRELT PAQIKKLITP RPNGKILFKR IIHGNRLVDI KQSAYGSKQV FPTDVSI KQ TNFYQGSIMS NEQKIRIENE IINLKNEYND RKSTLDALSN QKSGYRHELS ELASKNDDIN REAHQLNEIR KKYTMRKS T IETLREKLDQ LKREARKDVS QKIKDIDDQI QQLLLKQRHL LSKMASSMKS LKNCQKELIS TQILQFEAQN MDVSMNDVI GFFNEREADL KSQYEDKKKF VKEMRDTPEF QSWMREIRSY DQDTKEKLNK VAEKYEEEGN FNLSFVQDVL DKLESEIAMV NHDESAVTI LDQVTAELRE LEHTVPQQSK DLETIKAKLK EDHAVLEPKL DDIVSKISAR FARLFNNVGS AGAVRLEKPK D YAEWKIEI MVKFRDNAPL KKLDSHTQSG GERAVSTVLY MIALQEFTSA PFRVVDQINQ GMDSRNERIV HKAMVENACA EN TSQYFLI TPKLLTGLHY HEKMRIHCVM AGSWIPNPSE DPKMIHFGET SNYSFD UniProtKB: Structural maintenance of chromosomes protein 5 |
-Macromolecule #2: Structural maintenance of chromosomes protein 6
| Macromolecule | Name: Structural maintenance of chromosomes protein 6 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 128.198742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI ...String: MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR SSSDVATADQ DNFLEESPSG YIKKVILRN FMCHEHFELE LGSRLNFIVG NNGSGKSAIL TAITIGLGAK ASETNRGSSL KDLIREGCYS AKIILHLDNS K YGAYQQGI FGNEIIVERI IKRDGPASFS LRSENGKEIS NKKKDIQTVV DYFSVPVSNP MCFLSQDAAR SFLTASTSQD KY SHFMKGT LLQEITENLL YASAIHDSAQ ENMALHLENL KSLKAEYEDA KKLLRELNQT SDLNERKMLL QAKSLWIDVA HNT DACKNL ENEISGIQQK VDEVTEKIRN RQEKIERYTS DGTTIEAQID AKVIYVNEKD SEHQNARELL RDVKSRFEKE KSNQ AEAQS NIDQGRKKVD ALNKTIAHLE EELTKEMGGD KDQMRQELEQ LEKANEKLRE VNNSLVVSLQ DVKNEERDIQ HERES ELRT ISRSIQNKKV ELQNIAKGND TFLMNFDRNM DRLLRTIEQR KNEFETPAIG PLGSLVTIRK GFEKWTRSIQ RAISSS LNA FVVSNPKDNR LFRDIMRSCG IRSNIPIVTY CLSQFDYSKG RAHGNYPTIV DALEFSKPEI ECLFVDLSRI ERIVLIE DK NEARNFLQRN PVNVNMALSL RDRRSGFQLS GGYRLDTVTY QDKIRLKVNS SSDNGTQYLK DLIEQETKEL QNIRDRYE E KLSEVRSRLK EIDGRLKSTK NEMRKTNFRM TELKMNVGKV VDTGILNSKI NERKNQEQAI ASYEAAKEEL GLKIEQIAQ EAQPIKEQYD STKLALVEAQ DELQQLKEDI NSRQSKIQKY KDDTIYYEDK KKVYLENIKK IEVNVAALKE GIQRQIQNAC AFCSKERIE NVDLPDTQEE IKRELDKVSR MIQKAEKSLG LSQEEVIALF EKCRNKYKEG QKKYMEIDEA LNRLHNSLKA R DQNYKNAE KGTCFDADMD FRASLKVRKF SGNLSFIKDT KSLEIYILTT NDEKARNVDT LSGGEKSFSQ MALLLATWKP MR SRIIALD QFDVFMDQVN RKIGTTLIVK KLKDIARTQT IIITPQDIGK IADIDSSGVS IHRMRDPERQ NNSNFYN UniProtKB: Structural maintenance of chromosomes protein 6 |
-Macromolecule #3: E3 SUMO-protein ligase MMS21
| Macromolecule | Name: E3 SUMO-protein ligase MMS21 / type: protein_or_peptide / ID: 3 Details: Saccharomyces cerevisiae Nse2 fused to N-terminal HA-tag. Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Aminoacyltransferases |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 31.817775 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSYPYDVPD YAGSGALNDN PIPKSVPLHP KSGKYFHNLH ARDLSNIYQQ CYKQIDETIN QLVDSTSPST IGIEEQVADI TSTYKLLST YESESNSFDE HIKDLKKNFK QSSDACPQID LSTWDKYRTG ELTAPKLSEL YLNMPTPEPA TMVNNTDTLK I LKVLPYIW ...String: MGSYPYDVPD YAGSGALNDN PIPKSVPLHP KSGKYFHNLH ARDLSNIYQQ CYKQIDETIN QLVDSTSPST IGIEEQVADI TSTYKLLST YESESNSFDE HIKDLKKNFK QSSDACPQID LSTWDKYRTG ELTAPKLSEL YLNMPTPEPA TMVNNTDTLK I LKVLPYIW NDPTCVIPDL QNPADEDDLQ IEGGKIELTC PITCKPYEAP LISRKCNHVF DRDGIQNYLQ GYTTRDCPQA AC SQVVSMR DFVRDPIMEL RCKIAKMKES QEQDKRSSQA IDVL UniProtKB: E3 SUMO-protein ligase MMS21 |
-Macromolecule #4: Non-structural maintenance of chromosomes element 1
| Macromolecule | Name: Non-structural maintenance of chromosomes element 1 / type: protein_or_peptide / ID: 4 Details: Saccharomyces cerevisiae Nse1 fused to N-terminal TEV-cleavable His6-tag. Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 40.943113 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGRENLYFQ GHMEVHEEQV SAPVTGDATA KYLLQYILSA RGICHENALI LALMRLETDA STLNTEWSIQ QWVDKLNDY INAINVKLNL LGYKIIRINH GIGRNAVTLK AKQNFESFED NTAIRAHNND YAVLQSIVLP ESNRFFVYVN L ASTEETKL ...String: MGSSHHHHHH SSGRENLYFQ GHMEVHEEQV SAPVTGDATA KYLLQYILSA RGICHENALI LALMRLETDA STLNTEWSIQ QWVDKLNDY INAINVKLNL LGYKIIRINH GIGRNAVTLK AKQNFESFED NTAIRAHNND YAVLQSIVLP ESNRFFVYVN L ASTEETKL ATRFNQNEIE FMKWAIEQFM ISGETIVEGP ALETSIIVKE VNRILVAATG DSNLAKWRKF STFTVGSTNL FQ FQELTAT DIEDLLLRLC ELKWFYRTQE GKFGIDLRCI AELEEYLTSM YNLNTCQNCH KLAIQGVRCG NESCREENEE TGE NSLSQI WHVDCFKHYI THVSKNCDRC GSSLITEGVY VI UniProtKB: Non-structural maintenance of chromosomes element 1 |
-Macromolecule #5: Non-structural maintenance of chromosome element 3
| Macromolecule | Name: Non-structural maintenance of chromosome element 3 / type: protein_or_peptide / ID: 5 / Details: Saccharomyces cerevisiae Nse3, untagged. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 34.005531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSIDNDSDV DLTEDLAVAK IVKENPVARK MVRYILSRGE SQNSIITRNK LQSVIHEAAR EENIAKPSFS KMFMDINAIL YNVYGFELQ GLPSKNNMNA GGNGSNSNTN KSMPEPLGHR AQKFILLNNV PHSKNFDDFK ILQSAHTYEE LIVTGEYIGD D IASGTSNT ...String: MSSIDNDSDV DLTEDLAVAK IVKENPVARK MVRYILSRGE SQNSIITRNK LQSVIHEAAR EENIAKPSFS KMFMDINAIL YNVYGFELQ GLPSKNNMNA GGNGSNSNTN KSMPEPLGHR AQKFILLNNV PHSKNFDDFK ILQSAHTYEE LIVTGEYIGD D IASGTSNT LESKLSTDRD LVYKGVLSVI LCIVFFSKNN ILHQELIKFL ETFGIPSDGS KIAILNITIE DLIKSLEKRE YI VRLEEKS DTDGEVISYR IGRRTQAELG LESLEKLVQE IMGLEKEQTK SLHDDIIKSI GDSYSI UniProtKB: Non-structural maintenance of chromosome element 3 |
-Macromolecule #6: Non-structural maintenance of chromosome element 4
| Macromolecule | Name: Non-structural maintenance of chromosome element 4 / type: protein_or_peptide / ID: 6 Details: Saccharomyces cerevisiae Nse4 fused to a C-terminal HALO-myc tag. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 81.441133 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS ...String: MSSTVISRKR RNSTVTEPDS SGETRKQKKS RSDEKSSSSK DGDPQLEFKV LQGYRDLESE MHKGRAQVTR TGDIGVAMDN LNAVDSLFN KVIGIKNNGL FAHDARAMVS ISELAQISVR NLKFDDSRSM VNLENIVNSL KRYMLKEHFK LNNIAENRND L TLAADEQS AADQQEESDG DIDRTPDDNH TDKATSSFKA TSMRHSYLQQ FSHYNEFSQF NWFRIGALYN TISKNAPITD HL MGPLSIE KKPRVLTQRR RNNDQVGEKI TAEKITQHSL NSTQQETTPE QVKKCFKKLS KKLGPEGSIN LFKFIIDPNS FSR SIENLF YTSFLIKEGK LLMEHDEEGL PTIKIKQSIS HTDSRSKEIE RQRRRAAHQN HIIFQMDMPT WRKLIKKYNI TSPF LDGSS GMAEIGTGFP FDPHYVEVLG ERMHYVDVGP RDGTPVLFLH GNPTSSYVWR NIIPHVAPTH RCIAPDLIGM GKSDK PDLG YFFDDHVRFM DAFIEALGLE EVVLVIHDWG SALGFHWAKR NPERVKGIAF MEFIRPIPTW DEWPEFARET FQAFRT TDV GRKLIIDQNV FIEGTLPMGV VRPLTEVEMD HYREPFLNPV DREPLWRFPN ELPIAGEPAN IVALVEEYMD WLHQSPV PK LLFWGTPGVL IPPAEAARLA KSLPNCKAVD IGPGLNLLQE DNPDLIGSEI ARWLSTLEIS GGSEQKLISE EDL UniProtKB: Non-structural maintenance of chromosome element 4 |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. Details: Glow-discharged for a period of 60 seconds at 15mA, PELCO easiGlow, Leica Microsystems, Germany. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: LEICA EM GP Details: Held in chamber for a period of 10 seconds, before blotting for 2.5 to 4.5 seconds (auto-sensor) then plunged into liquid ethane before being stored under liquid nitrogen.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 4 / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: OTHER / Software - Name: cryoSPARC (ver. 3.1.0) / Number images used: 1 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC (ver. 3.1.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.1.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-7qcd: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)