[English] 日本語
 Yorodumi
Yorodumi- EMDB-13587: Photorhabdus laumondii T6SS-associated Rhs protein carrying the T... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13587 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Photorhabdus laumondii T6SS-associated Rhs protein carrying the Tre23 toxin domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RHS / TVISS / T6SS / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRHS repeat / RHS Repeat / Domain of unknown function DUF6531 / Domain of unknown function (DUF6531) / PAAR motif / PAAR motif / : / Teneurin YD-shell / YD repeat / : / Rhs repeat-associated core Similarity search - Domain/homology | |||||||||
| Biological species |  Photorhabdus laumondii (bacteria) / Photorhabdus laumondii (bacteria) /  Photorhabdus laumondii subsp. laumondii TTO1 (bacteria) Photorhabdus laumondii subsp. laumondii TTO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Jurenas D / Talachia Rosa L | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Mounting, structure and autocleavage of a type VI secretion-associated Rhs polymorphic toxin. Authors: Dukas Jurėnas / Leonardo Talachia Rosa / Martial Rey / Julia Chamot-Rooke / Rémi Fronzes / Eric Cascales /  Abstract: Bacteria have evolved toxins to outcompete other bacteria or to hijack host cell pathways. One broad family of bacterial polymorphic toxins gathers multidomain proteins with a modular organization, ...Bacteria have evolved toxins to outcompete other bacteria or to hijack host cell pathways. One broad family of bacterial polymorphic toxins gathers multidomain proteins with a modular organization, comprising a C-terminal toxin domain fused to a N-terminal domain that adapts to the delivery apparatus. Polymorphic toxins include bacteriocins, contact-dependent growth inhibition systems, and specialized Hcp, VgrG, PAAR or Rhs Type VI secretion (T6SS) components. We recently described and characterized Tre23, a toxin domain fused to a T6SS-associated Rhs protein in Photorhabdus laumondii, Rhs1. Here, we show that Rhs1 forms a complex with the T6SS spike protein VgrG and the EagR chaperone. Using truncation derivatives and cross-linking mass spectrometry, we demonstrate that VgrG-EagR-Rhs1 complex formation requires the VgrG C-terminal β-helix and the Rhs1 N-terminal region. We then report the cryo-electron-microscopy structure of the Rhs1-EagR complex, demonstrating that the Rhs1 central region forms a β-barrel cage-like structure that encapsulates the C-terminal toxin domain, and provide evidence for processing of the Rhs1 protein through aspartyl autoproteolysis. We propose a model for Rhs1 loading on the T6SS, transport and delivery into the target cell. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13587.map.gz emd_13587.map.gz | 65.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13587-v30.xml emd-13587-v30.xml emd-13587.xml emd-13587.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
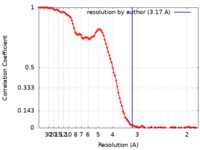
| FSC (resolution estimation) |  emd_13587_fsc.xml emd_13587_fsc.xml | 12.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13587.png emd_13587.png | 49.6 KB | ||
| Masks |  emd_13587_msk_1.map emd_13587_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13587.cif.gz emd-13587.cif.gz | 6.6 KB | ||
| Others |  emd_13587_half_map_1.map.gz emd_13587_half_map_1.map.gz emd_13587_half_map_2.map.gz emd_13587_half_map_2.map.gz | 65.5 MB 65.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13587 http://ftp.pdbj.org/pub/emdb/structures/EMD-13587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13587 | HTTPS FTP |
-Related structure data
| Related structure data |  7pq5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13587.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13587.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
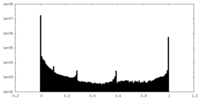
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
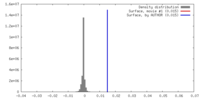
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13587_msk_1.map emd_13587_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
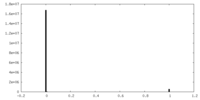
| Density Histograms |
-Half map: #2
| File | emd_13587_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
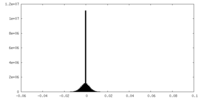
| Density Histograms |
-Half map: #1
| File | emd_13587_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tre23
| Entire | Name: Tre23 |
|---|---|
| Components |
|
-Supramolecule #1: Tre23
| Supramolecule | Name: Tre23 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Photorhabdus laumondii (bacteria) Photorhabdus laumondii (bacteria) |
-Macromolecule #1: Tre23
| Macromolecule | Name: Tre23 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Photorhabdus laumondii subsp. laumondii TTO1 (bacteria) Photorhabdus laumondii subsp. laumondii TTO1 (bacteria) |
| Molecular weight | Theoretical: 165.588828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLGDEIVTR IARVGARHAV HVSPPQGSPG PAAAREGDPI KHASFLGALA GAITGALIGA AVFAAASALV GLTILTGGLA TVAVIALGT AATFALGDVI SAASSAVTNM VDSIGPPSGT LTSGSPNVFI EGQPAARATT DIAVCSKHPA PPLIAQGSET V LINGSPAA ...String: MSLGDEIVTR IARVGARHAV HVSPPQGSPG PAAAREGDPI KHASFLGALA GAITGALIGA AVFAAASALV GLTILTGGLA TVAVIALGT AATFALGDVI SAASSAVTNM VDSIGPPSGT LTSGSPNVFI EGQPAARATT DIAVCSKHPA PPLIAQGSET V LINGSPAA RVDDKLACGA TIKSGAKTVF IGSGQGTYLA VADEFSAWER AILIAVEFLV PPSRGALKGL GKLFTKPGQG IQ GVLKGAK AGAKKAKALL SNRISCAKKA FRETEGAKRY REATKKFFTG DPIDVTTGQL FDQRTDITLG QTLPLVFLRS WVP EEQGLL GPGWTDSFSE CALATGDRVE IRTTEGASLY FALPAAYTHS VNPDHPDFTL SRGEQGYILR HRDSPVSKYF TLPH PSSAS KEAPRRWLLT EHRDVYDNRL RFIYNKHCQL TQVLHSDGPE LTLLYNLRGQ LTEIRRTDER LQEVMARYHY HDNGR LAEA DSTQNFHLYY EYNAQGLISR WSDGDQTWVD YRYDKQGRCT DSVGAGGFYP VHLDYAPGIT RSTTPQGHTT TGHYND QQL ITEIHTPCGG VTRYEYDRWG NLVRQILPEG ETLTLTYLAD TGRVTSLTEA TGAVWQYSYE ADSLQLTGMT DPLQRTW LP QYDEQGQPAG FIAPDGRKTT LTRNAFGLVT SETDPDGNSR TQEYDKHQRL VRVLDEENRT VSLGYDSQDR LRSLTAAG A LWRWRYDRHH RVAVSDRPDN QLEHFTHDRH GNLTCWTDAR GVKWQVEYGP FDLPVARRDG EGHRWQYRYD ADTLQLTQV INPQGETYSY TLDADGRVIT EQDYAGTQWH YRYDRSGNCI EKRDGEENVT RYDYDAARRL TTLHTPEGPT RYHYDSVGRL LTVDSPDST LHFEYDGQDR IVREIQPHGE IQRHYPDNRT AERQLLTGET APVTGPARFS GQTHPGRWQS RREVNRVGEL I TLTLAGQA PLTIERDDAG RDTGRYVDGG FILRQQYSLM GQLTAQRAGR NPYFFRPAEP DEIEGPAYAG VARRYEYDTA LN LTAASDD GQQVNYLLNG NGQVISVGEG RTLREHYQYD ETGYPSRRFD GVQEIMGETL YQEGHRLNWV GSHRFVYDRA GRM QEKQFL AEGCRLALTK YRWNSQNQLT GLITPDGIPW EYRYDAFGRR TEKRCIQSGK LTTYLWDGNV PAEIREYQHG RLKM IRHLV FDGWELVAQQ TQAFTLNLDN RVELMAGEVQ TQYAVSAPTG EPLALFDPAG KRVWRRPKQS LYGLRLGGYG ENPQL DPGL RFAGQLFDEE SGLFYNRFRY YLPEATCYLS PDPTGLWGGE NTYRYVQNPT KFINPLGLAG ENVFIHATNK AGFDGI MKS GVLHPNVSGK VAITDVLMSP KDVMRDLLIN NPNHIGRGDY AIIFKIDPGE RGNIRSPSRL EYTHEGKLKL NNILYAG KN PYAILEHLDY DTRLKLTDNQ VKIRKCGGKV DDYKDDDDK UniProtKB: Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 2/17 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8.5 Details: 50 mM Tris-HCl pH8.5, 250 mM NaCl, 1 mM TCEP, cOmpleteTM protease inhibitor cocktail (Sigma-Aldrich) |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 35 sec. / Pretreatment - Pressure: 25.0 kPa / Details: 2 mAu current |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-37 / Number grids imaged: 1 / Number real images: 8550 / Average exposure time: 3.7 sec. / Average electron dose: 49.75 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 20.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 45000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Overall B value: 75.76 |
|---|---|
| Output model |  PDB-7pq5: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)