[English] 日本語
 Yorodumi
Yorodumi- EMDB-13146: Human mitochondrial Lon protease with substrate in the ATPase domain -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13146 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial Lon protease with substrate in the ATPase domain | |||||||||
 Map data Map data | Composite map of human mitochondrial LonP1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protease / Mitochondria / AAA+ / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationoxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / mitochondrial protein catabolic process / endopeptidase La / G-quadruplex DNA binding / mitochondrial DNA metabolic process / : / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins ...oxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / mitochondrial protein catabolic process / endopeptidase La / G-quadruplex DNA binding / mitochondrial DNA metabolic process / : / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid / insulin receptor substrate binding / Mitochondrial unfolded protein response (UPRmt) / chaperone-mediated protein complex assembly / response to hormone / DNA polymerase binding / Mitochondrial protein degradation / negative regulation of insulin receptor signaling pathway / proteolysis involved in protein catabolic process / mitochondrion organization / protein catabolic process / ADP binding / single-stranded DNA binding / cellular response to oxidative stress / sequence-specific DNA binding / response to hypoxia / single-stranded RNA binding / mitochondrial matrix / serine-type endopeptidase activity / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Valentin Gese G / Shahzad S | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: A dual allosteric pathway drives human mitochondrial Lon Authors: Valentin Gese G / Shahzad S / Pardo-Hernandez C / Wramstedt A / Falkenberg M / Hallberg M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13146.map.gz emd_13146.map.gz | 43.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13146-v30.xml emd-13146-v30.xml emd-13146.xml emd-13146.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13146.png emd_13146.png | 97.9 KB | ||
| Filedesc metadata |  emd-13146.cif.gz emd-13146.cif.gz | 6 KB | ||
| Others |  emd_13146_additional_1.map.gz emd_13146_additional_1.map.gz emd_13146_additional_2.map.gz emd_13146_additional_2.map.gz emd_13146_additional_3.map.gz emd_13146_additional_3.map.gz | 778 MB 769.1 MB 756.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13146 http://ftp.pdbj.org/pub/emdb/structures/EMD-13146 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13146 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13146 | HTTPS FTP |
-Validation report
| Summary document |  emd_13146_validation.pdf.gz emd_13146_validation.pdf.gz | 368.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13146_full_validation.pdf.gz emd_13146_full_validation.pdf.gz | 368 KB | Display | |
| Data in XML |  emd_13146_validation.xml.gz emd_13146_validation.xml.gz | 8.6 KB | Display | |
| Data in CIF |  emd_13146_validation.cif.gz emd_13146_validation.cif.gz | 10 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13146 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13146 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13146 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13146 | HTTPS FTP |
-Related structure data
| Related structure data |  7p09MC  7p0bC  7p0mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13146.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13146.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of human mitochondrial LonP1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.654 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
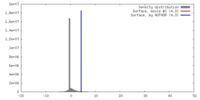
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Human mitochondrial LonP1 protease, ATPase domain focus
| File | emd_13146_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human mitochondrial LonP1 protease, ATPase domain focus | ||||||||||||
| Projections & Slices |
| ||||||||||||
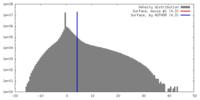
| Density Histograms |
-Additional map: Human mitochondrial LonP1 protease, Lan domain focus
| File | emd_13146_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human mitochondrial LonP1 protease, Lan domain focus | ||||||||||||
| Projections & Slices |
| ||||||||||||
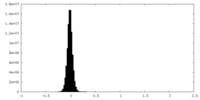
| Density Histograms |
-Additional map: Human mitochondrial LonP1 protease, protease domain focus
| File | emd_13146_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human mitochondrial LonP1 protease, protease domain focus | ||||||||||||
| Projections & Slices |
| ||||||||||||
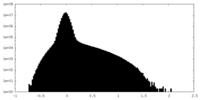
| Density Histograms |
- Sample components
Sample components
-Entire : Composite map of human mitochondrial LonP1
| Entire | Name: Composite map of human mitochondrial LonP1 |
|---|---|
| Components |
|
-Supramolecule #1: Composite map of human mitochondrial LonP1
| Supramolecule | Name: Composite map of human mitochondrial LonP1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Composite map of three maps refined with a focus on the Lan domains, the ATPase domains and the protease domains, respectively |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 591 KDa |
-Macromolecule #1: Lon protease homolog, mitochondrial
| Macromolecule | Name: Lon protease homolog, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.673164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMGFWEASSR GGGAFSGGED ASEGGAEEGA GGAGGSAGAG EGPVITALTP MTIPDVFPHL PLIAITRNPV FPRFIKIIEV KNKKLVELL RRKVRLAQPY VGVFLKRDDS NESDVVESLD EIYHTGTFAQ IHEMQDLGDK LRMIVMGHRR VHISRQLEVE P EEPEAENK ...String: SMGFWEASSR GGGAFSGGED ASEGGAEEGA GGAGGSAGAG EGPVITALTP MTIPDVFPHL PLIAITRNPV FPRFIKIIEV KNKKLVELL RRKVRLAQPY VGVFLKRDDS NESDVVESLD EIYHTGTFAQ IHEMQDLGDK LRMIVMGHRR VHISRQLEVE P EEPEAENK HKPRRKSKRG KKEAEDELSA RHPAELAMEP TPELPAEVLM VEVENVVHED FQVTEEVKAL TAEIVKTIRD II ALNPLYR ESVLQMMQAG QRVVDNPIYL SDMGAALTGA ESHELQDVLE ETNIPKRLYK ALSLLKKEFE LSKLQQRLGR EVE EKIKQT HRKYLLQEQL KIIKKELGLE KDDKDAIEEK FRERLKELVV PKHVMDVVDE ELSKLGLLDN HSSEFNVTRN YLDW LTSIP WGKYSNENLD LARAQAVLEE DHYGMEDVKK RILEFIAVSQ LRGSTQGKIL CFYGPPGVGK TSIARSIARA LNREY FRFS VGGMTDVAEI KGHRRTYVGA MPGKIIQCLK KTKTENPLIL IDEVDKIGRG YQGDPSSALL ELLDPEQNAN FLDHYL DVP VDLSKVLFIC TANVTDTIPE PLRDRMEMIN VSGYVAQEKL AIAERYLVPQ ARALCGLDES KAKLSSDVLT LLIKQYC RE SGVRNLQKQV EKVLRKSAYK IVSGEAESVE VTPENLQDFV GKPVFTVERM YDVTPPGVVM GLAWTAMGGS TLFVETSL R RPQDKDAKGD KDGSLEVTGQ LGEVMKESAR IAYTFARAFL MQHAPANDYL VTSHIHLHVP EGATPKDGPS AGCTIVTAL LSLAMGRPVR QNLAMTGEVS LTGKILPVGG IKEKTIAAKR AGVTCIVLPA ENKKDFYDLA AFITEGLEVH FVEHYREIFD IAFPD UniProtKB: Lon protease homolog, mitochondrial |
-Macromolecule #2: Unknown peptide from human mitochondrial transcription factor A (TFAM)
| Macromolecule | Name: Unknown peptide from human mitochondrial transcription factor A (TFAM) type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 954.168 Da |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 1.5 sec. / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF Details: This is a combined map from three focused refienements. The Lan domain, the ATPase domain and the protease domain focused maps with a 0.143 FSC resolution of 7.4, 2.7 and 2.75 angstroms. Number images used: 152455 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)