+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11966 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Puumala virus-like particle glycoprotein lattice. | |||||||||
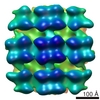
 Map data Map data | A reconstruction of a Puumala virus-like particle glycoprotein spike. | |||||||||
 Sample Sample | Puumala virus - Sotkamo != Puumala orthohantavirus Puumala virus - Sotkamo
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / host cell surface / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host innate immune response / virus-mediated perturbation of host defense response / virion membrane / cell surface / signal transduction ...symbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / host cell surface / host cell endoplasmic reticulum membrane / symbiont-mediated suppression of host innate immune response / virus-mediated perturbation of host defense response / virion membrane / cell surface / signal transduction / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Puumala orthohantavirus Puumala orthohantavirus | |||||||||
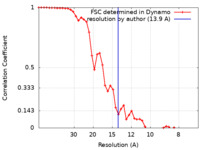
| Method | subtomogram averaging / cryo EM / Resolution: 13.9 Å | |||||||||
 Authors Authors | Rissanen I / Stass R / Huiskonen JT / Bowden TA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein. Authors: Ilona Rissanen / Robert Stass / Stefanie A Krumm / Jeffrey Seow / Ruben Jg Hulswit / Guido C Paesen / Jussi Hepojoki / Olli Vapalahti / Åke Lundkvist / Olivier Reynard / Viktor Volchkov / ...Authors: Ilona Rissanen / Robert Stass / Stefanie A Krumm / Jeffrey Seow / Ruben Jg Hulswit / Guido C Paesen / Jussi Hepojoki / Olli Vapalahti / Åke Lundkvist / Olivier Reynard / Viktor Volchkov / Katie J Doores / Juha T Huiskonen / Thomas A Bowden /      Abstract: The intricate lattice of Gn and Gc glycoprotein spike complexes on the hantavirus envelope facilitates host-cell entry and is the primary target of the neutralizing antibody-mediated immune response. ...The intricate lattice of Gn and Gc glycoprotein spike complexes on the hantavirus envelope facilitates host-cell entry and is the primary target of the neutralizing antibody-mediated immune response. Through study of a neutralizing monoclonal antibody termed mAb P-4G2, which neutralizes the zoonotic pathogen Puumala virus (PUUV), we provide a molecular-level basis for antibody-mediated targeting of the hantaviral glycoprotein lattice. Crystallographic analysis demonstrates that P-4G2 binds to a multi-domain site on PUUV Gc and may preclude fusogenic rearrangements of the glycoprotein that are required for host-cell entry. Furthermore, cryo-electron microscopy of PUUV-like particles in the presence of P-4G2 reveals a lattice-independent configuration of the Gc, demonstrating that P-4G2 perturbs the (Gn-Gc) lattice. This work provides a structure-based blueprint for rationalizing antibody-mediated targeting of hantaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11966.map.gz emd_11966.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11966-v30.xml emd-11966-v30.xml emd-11966.xml emd-11966.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11966_fsc.xml emd_11966_fsc.xml | 4.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11966.png emd_11966.png | 103.8 KB | ||
| Masks |  emd_11966_msk_1.map emd_11966_msk_1.map | 6.9 MB |  Mask map Mask map | |
| Others |  emd_11966_half_map_1.map.gz emd_11966_half_map_1.map.gz emd_11966_half_map_2.map.gz emd_11966_half_map_2.map.gz | 5.6 MB 5.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11966 http://ftp.pdbj.org/pub/emdb/structures/EMD-11966 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11966 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11966 | HTTPS FTP |
-Validation report
| Summary document |  emd_11966_validation.pdf.gz emd_11966_validation.pdf.gz | 448.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11966_full_validation.pdf.gz emd_11966_full_validation.pdf.gz | 447.2 KB | Display | |
| Data in XML |  emd_11966_validation.xml.gz emd_11966_validation.xml.gz | 10.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11966 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11966 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11966 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11966 | HTTPS FTP |
-Related structure data
| Related structure data |  7b0aMC  6z06C  7b09C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11966.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11966.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A reconstruction of a Puumala virus-like particle glycoprotein spike. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
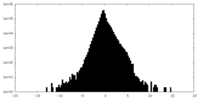
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11966_msk_1.map emd_11966_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
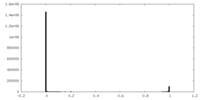
| Density Histograms |
-Half map: Half map (even) used to estimate resolution by FSC.
| File | emd_11966_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (even) used to estimate resolution by FSC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
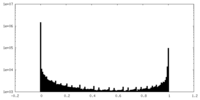
| Density Histograms |
-Half map: Half map (odd) used to estimate resolution by FSC.
| File | emd_11966_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (odd) used to estimate resolution by FSC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
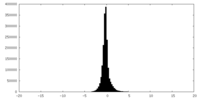
| Density Histograms |
- Sample components
Sample components
-Entire : Puumala virus - Sotkamo
| Entire | Name:  Puumala virus - Sotkamo Puumala virus - Sotkamo |
|---|---|
| Components |
|
-Supramolecule #1: Puumala orthohantavirus
| Supramolecule | Name: Puumala orthohantavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 1980486 / Sci species name: Puumala orthohantavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host system | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope polyprotein
| Macromolecule | Name: Envelope polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Puumala orthohantavirus Puumala orthohantavirus |
| Molecular weight | Theoretical: 49.827898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGETQNLNS GWTDTAHGSG IIPMRTDLEL DFSLPSSASY TYRRQLQNPA NEQEKIPFHL QISKQVIHAE IQHLGHWMDG TFNLKTAFH CYGSCEKYAY PWQTAGCFIE KDYEYESGWG CNPPDCPGVG TGCTACGVYL DKLKSVGKAF KIVSLRYTRK A CIQLGTEQ ...String: GPGETQNLNS GWTDTAHGSG IIPMRTDLEL DFSLPSSASY TYRRQLQNPA NEQEKIPFHL QISKQVIHAE IQHLGHWMDG TFNLKTAFH CYGSCEKYAY PWQTAGCFIE KDYEYESGWG CNPPDCPGVG TGCTACGVYL DKLKSVGKAF KIVSLRYTRK A CIQLGTEQ TCKSVDSNDC LVTTSVKVCL IGTVSKFQPS DTLLFLGPLE QGGLIFKQWC TTTCQFGDPG DIMSTPVGMK CP ELNGSFR KKCAFATTPV CQFDGNTLSG YKRMIATKDS FQSFNVTEPH ISASSLEWID PDSSLRDHIN VIVGRDLSFQ DLS ETPCQV DLATTSIDGA WGSGVGFNLV CSVSLTECST FLTSIKACDS AMCYGSTTAN LLRGQNTVHI VGKGGHSGSK FMCC HDTKC SSTGLVAAAP HLDRVTGYNQ ADSDKIFDDG APECGISCWF TKSGEGTETS QVAPA |
-Macromolecule #2: Envelope polyprotein
| Macromolecule | Name: Envelope polyprotein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Puumala orthohantavirus Puumala orthohantavirus |
| Molecular weight | Theoretical: 40.061566 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ETGELKIECP HTIGLGQGLV IGSVELPPVP LTQVESLKLE SSCNFDLHTS TSSQQPFTKW TWEMKSDLAE NTQASSTSFQ TKSSEINLR GLCLVPPLVI ETAARTRKTI ACFDLSCNQT ACQPTVFLIG PIQTCITTKS CLLGLGDQRI QVNYEKTYCV S GQLVEGVC ...String: ETGELKIECP HTIGLGQGLV IGSVELPPVP LTQVESLKLE SSCNFDLHTS TSSQQPFTKW TWEMKSDLAE NTQASSTSFQ TKSSEINLR GLCLVPPLVI ETAARTRKTI ACFDLSCNQT ACQPTVFLIG PIQTCITTKS CLLGLGDQRI QVNYEKTYCV S GQLVEGVC FNPVHTMALS QPSHTYDIVT VMVRCFLIAK KVSTGDSMKL EKSFETLVQK TSCTGNGFQG YYICLVGSSS EP LYIPTLD DYRSAEVLSR MAFAPHGEDH DVEKNAISAM RIIGKVTGKA PSTESSDTIQ GVAFSGNPLY TSTGVLTAKD DPV YIWAPG IIMEGNHSVC DKKTLPLTWT GFIPLPGEIE KTGTKHHHHH H |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 4.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)