+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Organisation of a helical Bunyamwera virus ribonucleoprotein | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | helix / ribonucleoprotein / nucleocapsid / virus / VIRAL PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報helical viral capsid / symbiont-mediated suppression of host gene expression / viral nucleocapsid / ribonucleoprotein complex / RNA binding 類似検索 - 分子機能 | |||||||||
| 生物種 |  Bunyamwera virus (ブニヤンベラウイルス) Bunyamwera virus (ブニヤンベラウイルス) | |||||||||
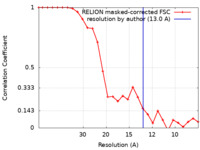
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 13.0 Å | |||||||||
 データ登録者 データ登録者 | Hopkins FR / Fontana J | |||||||||
| 資金援助 |  英国, 1件 英国, 1件
| |||||||||
 引用 引用 |  ジャーナル: mBio / 年: 2022 ジャーナル: mBio / 年: 2022タイトル: The Native Orthobunyavirus Ribonucleoprotein Possesses a Helical Architecture. 著者: Francis R Hopkins / Beatriz Álvarez-Rodríguez / George R Heath / Kyriakoulla Panayi / Samantha Hover / Thomas A Edwards / John N Barr / Juan Fontana /  要旨: The order is the largest group of negative-sense RNA viruses, containing many lethal human pathogens for which approved anti-infective measures are not available. The bunyavirus genome consists of ...The order is the largest group of negative-sense RNA viruses, containing many lethal human pathogens for which approved anti-infective measures are not available. The bunyavirus genome consists of multiple negative-sense RNA segments enwrapped by the virus-encoded nucleocapsid protein (NP), which together with the viral polymerase form ribonucleoproteins (RNPs). RNPs represent substrates for RNA synthesis and virion assembly, which require inherent flexibility, consistent with the appearance of RNPs spilled from virions. These observations have resulted in conflicting models describing the overall RNP architecture. Here, we purified RNPs from Bunyamwera virus (BUNV), the prototypical orthobunyavirus. The lengths of purified RNPs imaged by negative staining resulted in 3 populations of RNPs, suggesting that RNPs possess a consistent method of condensation. Employing microscopy approaches, we conclusively show that the NP portion of BUNV RNPs is helical. Furthermore, we present a pseudo-atomic model for this portion based on a cryo-electron microscopy average at 13 Å resolution, which allowed us to fit the BUNV NP crystal structure by molecular dynamics. This model was confirmed by NP mutagenesis using a mini-genome system. The model shows that adjacent NP monomers in the RNP chain interact laterally through flexible N- and C-terminal arms only, with no longitudinal helix-stabilizing interactions, thus providing a potential model for the molecular basis for RNP flexibility. Excessive RNase treatment disrupts native RNPs, suggesting that RNA was key in maintaining the RNP structure. Overall, this work will inform studies on bunyaviral RNP assembly, packaging, and RNA replication, and aid in future antiviral strategies. Bunyaviruses are emerging RNA viruses that cause significant disease and economic burden and for which vaccines or therapies approved for humans are not available. The bunyavirus genome is wrapped up by the nucleoprotein (NP) and interacts with the viral polymerase, forming a ribonucleoprotein (RNP). This is the only form of the genome active for viral replication and assembly. However, until now how NPs are organized within an RNP was not known for any orthobunyavirus. Here, we purified RNPs from the prototypical orthobunyavirus, Bunyamwera virus, and employed microscopy approaches to show that the NP portion of the RNP was helical. We then combined our helical average with the known structure of an NP monomer, generating a pseudo-atomic model of this region. This arrangement allowed the RNPs to be highly flexible, which was critical for several stages of the viral replication cycle, such as segment circularization. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_11847.map.gz emd_11847.map.gz | 735.8 KB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-11847-v30.xml emd-11847-v30.xml emd-11847.xml emd-11847.xml | 9.9 KB 9.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_11847_fsc.xml emd_11847_fsc.xml | 2.4 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_11847.png emd_11847.png | 24.8 KB | ||
| Filedesc metadata |  emd-11847.cif.gz emd-11847.cif.gz | 5.1 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11847 http://ftp.pdbj.org/pub/emdb/structures/EMD-11847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11847 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11847 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_11847_validation.pdf.gz emd_11847_validation.pdf.gz | 425.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_11847_full_validation.pdf.gz emd_11847_full_validation.pdf.gz | 424.6 KB | 表示 | |
| XML形式データ |  emd_11847_validation.xml.gz emd_11847_validation.xml.gz | 6.5 KB | 表示 | |
| CIF形式データ |  emd_11847_validation.cif.gz emd_11847_validation.cif.gz | 7.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11847 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11847 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11847 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7aoyMC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_11847.map.gz / 形式: CCP4 / 大きさ: 1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_11847.map.gz / 形式: CCP4 / 大きさ: 1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 4.28 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Native ribonucleoprotein extracted from Bunyamwera virus
| 全体 | 名称: Native ribonucleoprotein extracted from Bunyamwera virus |
|---|---|
| 要素 |
|
-超分子 #1: Native ribonucleoprotein extracted from Bunyamwera virus
| 超分子 | 名称: Native ribonucleoprotein extracted from Bunyamwera virus タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Bunyamwera virus (ブニヤンベラウイルス) Bunyamwera virus (ブニヤンベラウイルス) |
-分子 #1: Nucleoprotein
| 分子 | 名称: Nucleoprotein / タイプ: protein_or_peptide / ID: 1 / コピー数: 9 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Bunyamwera virus (ブニヤンベラウイルス) Bunyamwera virus (ブニヤンベラウイルス) |
| 分子量 | 理論値: 26.69876 KDa |
| 配列 | 文字列: MIELEFHDVA ANTSSTFDPE VAYANFKRVH TTGLSYDHIR IFYIKGREIK TSLAKRSEWE VTLNLGGWKI TVYNTNFPGN RNNPVPDDG LTLHRLSGFL ARYLLEKMLK VSEPEKLIIK SKIINPLAEK NGITWNDGEE VYLSFFPGSE MFLGTFRFYP L AIGIYKVQ ...文字列: MIELEFHDVA ANTSSTFDPE VAYANFKRVH TTGLSYDHIR IFYIKGREIK TSLAKRSEWE VTLNLGGWKI TVYNTNFPGN RNNPVPDDG LTLHRLSGFL ARYLLEKMLK VSEPEKLIIK SKIINPLAEK NGITWNDGEE VYLSFFPGSE MFLGTFRFYP L AIGIYKVQ RKEMEPKYLE KTMRQRYMGL EAATWTVSKL TEVQSALTVV SSLGWKKTNV SAAARDFLAK FGINM UniProtKB: Nucleoprotein |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | filament |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| 凍結 | 凍結剤: NITROGEN / 装置: FEI VITROBOT MARK II |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | 位相板: VOLTA PHASE PLATE |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 平均露光時間: 9.0 sec. / 平均電子線量: 51.3 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 倍率(公称値): 130000 |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)