+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10203 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
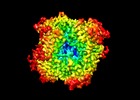
| Title | Escherichia coli AGPase in complex with AMP. Symmetry C2 | |||||||||
 Map data Map data | Sharp map of the full map of the single-particle reconstruction of Escherichia coli AGPase in complex with AMP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ADP-glucose pyrophosphorilase Complex with AMP inhibitor / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucose-1-phosphate adenylyltransferase complex / glucose-1-phosphate adenylyltransferase / glucose-1-phosphate adenylyltransferase activity / glycogen biosynthetic process / AMP binding / protein homotetramerization / magnesium ion binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
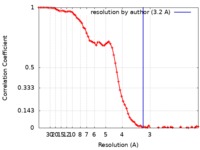
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Cifuente JO / Comino N | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Res Struct Biol / Year: 2020 Journal: Curr Res Struct Biol / Year: 2020Title: The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM. Authors: Javier O Cifuente / Natalia Comino / Cecilia D'Angelo / Alberto Marina / David Gil-Carton / David Albesa-Jové / Marcelo E Guerin /  Abstract: Glycogen and starch are the major carbon and energy reserve polysaccharides in nature, providing living organisms with a survival advantage. The evolution of the enzymatic machinery responsible for ...Glycogen and starch are the major carbon and energy reserve polysaccharides in nature, providing living organisms with a survival advantage. The evolution of the enzymatic machinery responsible for the biosynthesis and degradation of such polysaccharides, led the development of mechanisms to control the assembly and disassembly rate, to store and recover glucose according to cell energy demands. The tetrameric enzyme ADP-glucose pyrophosphorylase (AGPase) catalyzes and regulates the initial step in the biosynthesis of both α-polyglucans. AGPase displays cooperativity and allosteric regulation by sensing metabolites from the cell energy flux. The understanding of the allosteric signal transduction mechanisms in AGPase arises as a long-standing challenge. In this work, we disclose the cryoEM structures of the paradigmatic homotetrameric AGPase from (AGPase), in complex with either positive or negative physiological allosteric regulators, fructose-1,6-bisphosphate (FBP) and AMP respectively, both at 3.0 Å resolution. Strikingly, the structures reveal that FBP binds deeply into the allosteric cleft and overlaps the AMP site. As a consequence, FBP promotes a concerted conformational switch of a regulatory loop, RL2, from a "locked" to a "free" state, modulating ATP binding and activating the enzyme. This notion is strongly supported by our complementary biophysical and bioinformatics evidence, and a careful analysis of vast enzyme kinetics data on single-point mutants of AGPase. The cryoEM structures uncover the residue interaction networks (RIN) between the allosteric and the catalytic components of the enzyme, providing unique details on how the signaling information is transmitted across the tetramer, from which cooperativity emerges. Altogether, the conformational states visualized by cryoEM reveal the regulatory mechanism of AGPase, laying the foundations to understand the allosteric control of bacterial glycogen biosynthesis at the molecular level of detail. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM Authors: Cifuente JO / Comino N / D'Angelo C / Marina A / Gil-Carton D / Albesa-Jove D / Guerin ME | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10203.map.gz emd_10203.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10203-v30.xml emd-10203-v30.xml emd-10203.xml emd-10203.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10203_fsc.xml emd_10203_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10203.png emd_10203.png | 150.8 KB | ||
| Filedesc metadata |  emd-10203.cif.gz emd-10203.cif.gz | 5.9 KB | ||
| Others |  emd_10203_additional_1.map.gz emd_10203_additional_1.map.gz emd_10203_additional_2.map.gz emd_10203_additional_2.map.gz emd_10203_additional_3.map.gz emd_10203_additional_3.map.gz | 28.2 MB 28.2 MB 15.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10203 http://ftp.pdbj.org/pub/emdb/structures/EMD-10203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10203 | HTTPS FTP |
-Related structure data
| Related structure data |  6shqMC  6shjC  6shnC  6si8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10203.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10203.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp map of the full map of the single-particle reconstruction of Escherichia coli AGPase in complex with AMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: First half map of the single-particle reconstruction of...
| File | emd_10203_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map of the single-particle reconstruction of Escherichia coli AGPase in complex with AMP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Second half map of the single-particle reconstruction of...
| File | emd_10203_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map of the single-particle reconstruction of Escherichia coli AGPase in complex with AMP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Full map of the single-particle reconstruction of Escherichia...
| File | emd_10203_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map of the single-particle reconstruction of Escherichia coli AGPase in complex with AMP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ADP.glucose pyrophosphorylase in complex with the inhibitor AMP
| Entire | Name: ADP.glucose pyrophosphorylase in complex with the inhibitor AMP |
|---|---|
| Components |
|
-Supramolecule #1: ADP.glucose pyrophosphorylase in complex with the inhibitor AMP
| Supramolecule | Name: ADP.glucose pyrophosphorylase in complex with the inhibitor AMP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Homotetrameric enzyme |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 194 KDa |
-Macromolecule #1: Glucose-1-phosphate adenylyltransferase
| Macromolecule | Name: Glucose-1-phosphate adenylyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glucose-1-phosphate adenylyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.75859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVSLEKNDHL MLARQLPLKS VALILAGGRG TRLKDLTNKR AKPAVHFGGK FRIIDFALSN CINSGIRRMG VITQYQSHTL VQHIQRGWS FFNEEMNEFV DLLPAQQRMK GENWYRGTAD AVTQNLDIIR RYKAEYVVIL AGDHIYKQDY SRMLIDHVEK G ARCTVACM ...String: MVSLEKNDHL MLARQLPLKS VALILAGGRG TRLKDLTNKR AKPAVHFGGK FRIIDFALSN CINSGIRRMG VITQYQSHTL VQHIQRGWS FFNEEMNEFV DLLPAQQRMK GENWYRGTAD AVTQNLDIIR RYKAEYVVIL AGDHIYKQDY SRMLIDHVEK G ARCTVACM PVPIEEASAF GVMAVDENDK IIEFVEKPAN PPSMPNDPSK SLASMGIYVF DADYLYELLE EDDRDENSSH DF GKDLIPK ITEAGLAYAH PFPLSCVQSD PDAEPYWRDV GTLEAYWKAN LDLASVVPEL DMYDRNWPIR TYNESLPPAK FVQ DRSGSH GMTLNSLVSG GCVISGSVVV QSVLFSRVRV NSFCNIDSAV LLPEVWVGRS CRLRRCVIDR ACVIPEGMVI GENA EEDAR RFYRSEEGIV LVTREMLRKL GHKQER UniProtKB: Glucose-1-phosphate adenylyltransferase |
-Macromolecule #2: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK II | ||||||||
| Details | Sample with single particles and some linear chains of particles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Max: 80.0 K |
| Details | Titan Krios I - Ebic - Diamond Light Source |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)