[English] 日本語
 Yorodumi
Yorodumi- EMDB-0710: Dimeric structure of Machupo virus polymerase bound to vRNA promoter -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0710 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimeric structure of Machupo virus polymerase bound to vRNA promoter | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polymerase / RNA virus / VIRAL PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA replication / cap snatching / virion component / host cell cytoplasm / Hydrolases; Acting on ester bonds / hydrolase activity / RNA-directed RNA polymerase / RNA-dependent RNA polymerase activity / nucleotide binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Machupo mammarenavirus / Machupo mammarenavirus /  Machupo virus / synthetic construct (others) Machupo virus / synthetic construct (others) | |||||||||
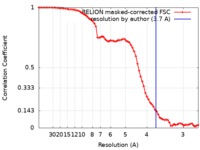
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Peng R / Xu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structural insight into arenavirus replication machinery. Authors: Ruchao Peng / Xin Xu / Jiamei Jing / Min Wang / Qi Peng / Sheng Liu / Ying Wu / Xichen Bao / Peiyi Wang / Jianxun Qi / George F Gao / Yi Shi /  Abstract: Arenaviruses can cause severe haemorrhagic fever and neurological diseases in humans and other animals, exemplified by Lassa mammarenavirus, Machupo mammarenavirus and lymphocytic choriomeningitis ...Arenaviruses can cause severe haemorrhagic fever and neurological diseases in humans and other animals, exemplified by Lassa mammarenavirus, Machupo mammarenavirus and lymphocytic choriomeningitis virus, posing great threats to public health. These viruses encode a large multi-domain RNA-dependent RNA polymerase for transcription and replication of the viral genome. Viral polymerases are one of the leading antiviral therapeutic targets. However, the structure of arenavirus polymerase is not yet known. Here we report the near-atomic resolution structures of Lassa and Machupo virus polymerases in both apo and promoter-bound forms. These structures display a similar overall architecture to influenza virus and bunyavirus polymerases but possess unique local features, including an arenavirus-specific insertion domain that regulates the polymerase activity. Notably, the ordered active site of arenavirus polymerase is inherently switched on, without the requirement for allosteric activation by 5'-viral RNA, which is a necessity for both influenza virus and bunyavirus polymerases. Moreover, dimerization could facilitate the polymerase activity. These findings advance our understanding of the mechanism of arenavirus replication and provide an important basis for developing antiviral therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0710.map.gz emd_0710.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0710-v30.xml emd-0710-v30.xml emd-0710.xml emd-0710.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0710_fsc.xml emd_0710_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0710.png emd_0710.png | 88.4 KB | ||
| Filedesc metadata |  emd-0710.cif.gz emd-0710.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0710 http://ftp.pdbj.org/pub/emdb/structures/EMD-0710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0710 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0710 | HTTPS FTP |
-Validation report
| Summary document |  emd_0710_validation.pdf.gz emd_0710_validation.pdf.gz | 440.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0710_full_validation.pdf.gz emd_0710_full_validation.pdf.gz | 440 KB | Display | |
| Data in XML |  emd_0710_validation.xml.gz emd_0710_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  emd_0710_validation.cif.gz emd_0710_validation.cif.gz | 12.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0710 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0710 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0710 | HTTPS FTP |
-Related structure data
| Related structure data |  6klhMC  0706C  0707C  0708C  0709C  6klcC  6kldC  6kleC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0710.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0710.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Machupo virus polymerase bound to vRNA promoter
| Entire | Name: Machupo virus polymerase bound to vRNA promoter |
|---|---|
| Components |
|
-Supramolecule #1: Machupo virus polymerase bound to vRNA promoter
| Supramolecule | Name: Machupo virus polymerase bound to vRNA promoter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Machupo mammarenavirus Machupo mammarenavirus |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Machupo virus Machupo virus |
| Molecular weight | Theoretical: 250.416062 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MDEYVQELKG LIRKHIPERC EFGHQKVTFL SQVHPSPLLT EGFKLLSSLV ELESCEAHAC QANTDQRFVD VILSDNGILC PTLPKVIPD GFKLTGKTLI LLETFVRVNP DEFEKKWKAD MSKLLNLKHD LQKSGVTLVP IVDGRSNYNN RFVADWVIER I RWLLIEIL ...String: MDEYVQELKG LIRKHIPERC EFGHQKVTFL SQVHPSPLLT EGFKLLSSLV ELESCEAHAC QANTDQRFVD VILSDNGILC PTLPKVIPD GFKLTGKTLI LLETFVRVNP DEFEKKWKAD MSKLLNLKHD LQKSGVTLVP IVDGRSNYNN RFVADWVIER I RWLLIEIL KASKSMLEID IEDQEYQRLI HSLSNVKNQS LGLENLEHLK RNSLDYDERL NESLFIGLKG DIRESTVREE LI KLKLWFK DEVFSKGLGK FKLTDRRELL ESLSSLGAHL DSDVSSCPFC NNKLMEIVYN VTFSCVERTD GVATVDQQFS TTH SNIEKH YLSVLSLCNK IKGLKVFNTR RNTLLFLDLI MVNLMVDISD SCQDAIESLR KSGLIVGQMV MLVNDRVLDI LEAV KLIRK KIGTNPNWVK NCSKILERSH PEIWHHLSTL IKQPDFNSLI SIAQHLVSDR PIMRYSVERG SDKICRHKLF QEMSS FEQM RLFKTLSSIS LSLINSMKTS FSSRLLVNER EFSKYFGNVR LRECYAQRFY LAESLVGFLF YQKTGERSRC YSVYLS DNG VMSEQGSFYC DPKRFFLPVF SDEVLAGMCE EMTSWLDFDT GLMNDTGPIL RLLVLAILCS PSKRNQTFLQ GLRYFLM AF ANQIHHIDLT SKLVVECKSS SEVVVQRLAV GLFIRLLSGE SDASLFFSRR FKYLLNVSYL CHLITKETPD RLTDQIKC F EKFIEPKVKF GCAVVNPSLN GKLTVDQEDI MINGLKKFFS KSLRDTEDVQ TPGVCKELLN YCVSLFNRGK LKVSGELKN NPFRPNITST ALDLSSNKSV VIPKLDELGN ILSTYDKEKL VSACVSSMAE RFKTKGRYNL DPDSTDYLIL KNLTGLVSAG PKAKSTQEE LSLMYEALTE EQVESFNEIK HDVQVALAKM ADNSVNTRTK NLGRADNSVK NGNNPLDNLW SPFGVMKEIR A EVSLHEVK DFDPDVLPPE VYKELCDAVY KSSEKCNFFL EGVLDVCPLG LLLKNLTTSS YVDEEYFMCF KYLLIQGHFD QK LGSYEHK SRSRLGFTDE TLRLKDEVRL SIRESNSEAI ADKLDKSYFT NAALRNLCFY SEDSPTEFTS ISSNSGNLKF GLS YKEQVG SNRELYVGDL NTKLMTRLVE DFSEAVGNSM KYTCLNSEKE FERAICDMKM AVNNGDLSCS YDHSKWGPTM SPAL FLALL QMLELRTPVD RSKIDLDSVK SILKWHLHKV VEVPINVAEA YCIGKLKRSL GLMGCGSTSL SEEFFHQTMQ LNGQI PSHI MSVLDMGQGI LHNTSDLYGL ITEQFLCYAL DLLYDVIPVS YTSSDDQITL IKTPSLDIEG GSDAAEWLEM ICFHEF LSS KLNKFVSPKS VIGTFVAEFK SRFFVMGEET PLLTKFVAAA LHNVKCKTPT QLSETIDTIC DQCIANGVST KIVTRIS KR VNQLIRYSGY GETPFGAIED QDVKDWVDGS RGYRLQRKIE AIFHDDKETS FIRNCARKVF NDIKRGRIFE ENLINLIG R GGDEALTGFL QYAGCSEQEV NRVLNYRWVN LSSFGDLRLV LRTKLMTSRR VLEREEVPTL IKTLQSKLSR NFTKGVKKI LAESINKSAF QSSVASGFIG FCKSMGSKCV RDGKGGFLYI KEVYSGVSAC TCEICALKPK IIYCNNSLNK VSQFSKPILW DYFSLVLTN ACELGEWVFS TVKEPQKPLV LNNQNFFWAV KPKVVRQIED QLGMNHVLQS IRRNYPVLFD EHLTPFMNDL Q VSRTMDSG RLKFLDVCIA LDMMNENLGI ISHLLKTRDN SVYIVKQSDC ALAHIRQSSY TDWELGLSPQ QICTNFKTQL VL SSMVNPL VLSTSCLKSF FWFNEVLELE DDSQIELAEL TDFALMVKNQ NVSRAMFVED IAMGYVVSNF EGVRISLSNV MVD GVQLPP QEKAPDIGEL FGLKAENVIV GLVVQIDHVR MSTKFKLKRK MVYSFSLECI MDVGEIQNKE VILKVVAVDQ SVSG SGGNH MLLDGVSVVA SLPLFTGQAS FDLAAMLIES NLAGSNDNFL MRNVTLDLGG FSPELSDKYS YRLSGPENQE DPLVL KDGA FYVGGERLST YKVEFTGDLV VKALGALEDD ESVVSMLHQL WPYLKATSQV ILFQQEDFTI VHDLYKKQLT KSIESF GEW IEFTNFKVAY SKSLKELVIS DTQGSFRLKG VMCRPLASTP QVEDIE UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*...
| Macromolecule | Name: RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3') type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.069649 KDa |
| Sequence | String: GCCUAGGAUC CACUGUGCG |
-Macromolecule #3: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)