[English] 日本語
 Yorodumi
Yorodumi- EMDB-50422: SARS-CoV-2 (wuhan variant) Spike protein, with two RBD up and one... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
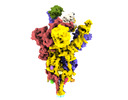
| Title | SARS-CoV-2 (wuhan variant) Spike protein, with two RBD up and one RBD down. Single chain fragment scFv41N bound to one RDB up and the RBD down | |||||||||
 Map data Map data | unsharped map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Spike / single chain fragment / VIRAL PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Berlinguer M / Chaves-Sanjuan A / Milazzo FM / Minenkova O / De Santis R / Bolognesi M | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of scFv41N in complex with SARS-CoV-2 Spike protein Authors: Chaves-Sanjuan A / Bolognesi M | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50422.map.gz emd_50422.map.gz | 121.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50422-v30.xml emd-50422-v30.xml emd-50422.xml emd-50422.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50422.png emd_50422.png | 61.1 KB | ||
| Masks |  emd_50422_msk_1.map emd_50422_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50422.cif.gz emd-50422.cif.gz | 6.2 KB | ||
| Others |  emd_50422_additional_1.map.gz emd_50422_additional_1.map.gz emd_50422_half_map_1.map.gz emd_50422_half_map_1.map.gz emd_50422_half_map_2.map.gz emd_50422_half_map_2.map.gz | 230.3 MB 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50422 http://ftp.pdbj.org/pub/emdb/structures/EMD-50422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50422 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50422 | HTTPS FTP |
-Validation report
| Summary document |  emd_50422_validation.pdf.gz emd_50422_validation.pdf.gz | 913.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50422_full_validation.pdf.gz emd_50422_full_validation.pdf.gz | 912.8 KB | Display | |
| Data in XML |  emd_50422_validation.xml.gz emd_50422_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_50422_validation.cif.gz emd_50422_validation.cif.gz | 19.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50422 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50422 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50422 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50422 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50422.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50422.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharped map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
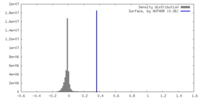
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50422_msk_1.map emd_50422_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
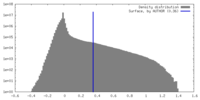
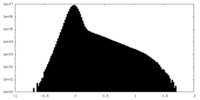
| Density Histograms |
-Additional map: sharped map
| File | emd_50422_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharped map | ||||||||||||
| Projections & Slices |
| ||||||||||||
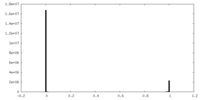
| Density Histograms |
-Half map: #1
| File | emd_50422_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
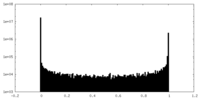
| Density Histograms |
-Half map: #2
| File | emd_50422_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Spike protein in complex with the single chain fragmen...
| Entire | Name: SARS-CoV-2 Spike protein in complex with the single chain fragment scFv41N |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Spike protein in complex with the single chain fragmen...
| Supramolecule | Name: SARS-CoV-2 Spike protein in complex with the single chain fragment scFv41N type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 510 KDa |
-Supramolecule #2: SARS-CoV-2 Spike protein
| Supramolecule | Name: SARS-CoV-2 Spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: scFv41N single chain fragment
| Supramolecule | Name: scFv41N single chain fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SARS-CoV-2 Spike protein
| Macromolecule | Name: SARS-CoV-2 Spike protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TPSALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGGGSGGSA WSHPQFEK |
-Macromolecule #2: scFv41N single chain fragment
| Macromolecule | Name: scFv41N single chain fragment / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RTMEEVQLLQ SAGGLVQPG G SLRLSCAA SG LTVSANY MSW VRQAPG KGLE WVSVI YPGGS TFYA DSVKGR FTI SRGNSKN TL YLQMNSLS V EDTAVYYCA RDLSVAGAFD IWGQGTLVT V SSGGGGSG GG GSGGGGS EIV LTQSPG TLSL SAGET ...String: RTMEEVQLLQ SAGGLVQPG G SLRLSCAA SG LTVSANY MSW VRQAPG KGLE WVSVI YPGGS TFYA DSVKGR FTI SRGNSKN TL YLQMNSLS V EDTAVYYCA RDLSVAGAFD IWGQGTLVT V SSGGGGSG GG GSGGGGS EIV LTQSPG TLSL SAGET ATLSC RASQ SISTDY LAW YQQKPGQ AP RLLIYGAS N RAAGIPDRF SGSASGTDFT LTISRLEAE D LAVYYCQQ YG SSPYTFG QGT KVEIKR AAAG DYKDD DDKHH HHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 6232 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)