+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HCoV-NL63 5' proximal stem-loop 5, conformation 1 | ||||||||||||||||||||||||||||||||||||
 Map data Map data | Conformation 1 of the HCoV-NL63 RNA SL5 domain. | ||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | SL5 / coronavirus / 5' UTR / RNA | ||||||||||||||||||||||||||||||||||||
| Biological species |  Human coronavirus NL63 Human coronavirus NL63 | ||||||||||||||||||||||||||||||||||||
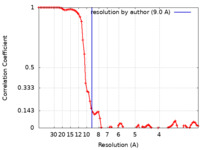
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Kretsch RC / Xu L / Zheludev IN / Zhou X / Huang R / Nye G / Li S / Zhang K / Chiu W / Das R | ||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 11 items United States, 11 items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses. Authors: Rachael C Kretsch / Lily Xu / Ivan N Zheludev / Xueting Zhou / Rui Huang / Grace Nye / Shanshan Li / Kaiming Zhang / Wah Chiu / Rhiju Das /   Abstract: Coronavirus genomes sequester their start codons within stem-loop 5 (SL5), a structured, 5' genomic RNA element. In most alpha- and betacoronaviruses, the secondary structure of SL5 is predicted to ...Coronavirus genomes sequester their start codons within stem-loop 5 (SL5), a structured, 5' genomic RNA element. In most alpha- and betacoronaviruses, the secondary structure of SL5 is predicted to contain a four-way junction of helical stems, some of which are capped with UUYYGU hexaloops. Here, using cryogenic electron microscopy (cryo-EM) and computational modeling with biochemically-determined secondary structures, we present three-dimensional structures of SL5 from six coronaviruses. The SL5 domain of betacoronavirus SARS-CoV-2, resolved at 4.7 Å resolution, exhibits a T-shaped structure, with its UUYYGU hexaloops at opposing ends of a coaxial stack, the T's "arms." Further analysis of SL5 domains from SARS-CoV-1 and MERS (7.1 and 6.4-6.9 Å resolution, respectively) indicate that the junction geometry and inter-hexaloop distances are conserved features across the studied human-infecting betacoronaviruses. The MERS SL5 domain displays an additional tertiary interaction, which is also observed in the non-human-infecting betacoronavirus BtCoV-HKU5 (5.9-8.0 Å resolution). SL5s from human-infecting alphacoronaviruses, HCoV-229E and HCoV-NL63 (6.5 and 8.4-9.0 Å resolution, respectively), exhibit the same coaxial stacks, including the UUYYGU-capped arms, but with a phylogenetically distinct crossing angle, an X-shape. As such, all SL5 domains studied herein fold into stable tertiary structures with cross-genus similarities, with implications for potential protein-binding modes and therapeutic targets. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42813.map.gz emd_42813.map.gz | 23.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42813-v30.xml emd-42813-v30.xml emd-42813.xml emd-42813.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42813_fsc.xml emd_42813_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42813.png emd_42813.png | 31.2 KB | ||
| Masks |  emd_42813_msk_1.map emd_42813_msk_1.map | 46.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42813.cif.gz emd-42813.cif.gz | 5.7 KB | ||
| Others |  emd_42813_half_map_1.map.gz emd_42813_half_map_1.map.gz emd_42813_half_map_2.map.gz emd_42813_half_map_2.map.gz | 43.1 MB 43.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42813 http://ftp.pdbj.org/pub/emdb/structures/EMD-42813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42813 | HTTPS FTP |
-Validation report
| Summary document |  emd_42813_validation.pdf.gz emd_42813_validation.pdf.gz | 652.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42813_full_validation.pdf.gz emd_42813_full_validation.pdf.gz | 651.9 KB | Display | |
| Data in XML |  emd_42813_validation.xml.gz emd_42813_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_42813_validation.cif.gz emd_42813_validation.cif.gz | 19.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42813 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42813 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42813.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42813.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Conformation 1 of the HCoV-NL63 RNA SL5 domain. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.482 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42813_msk_1.map emd_42813_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
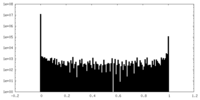
| Density Histograms |
-Half map: Half map of conformation 1 of the HCoV-NL63 RNA SL5 domain.
| File | emd_42813_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of conformation 1 of the HCoV-NL63 RNA SL5 domain. | ||||||||||||
| Projections & Slices |
| ||||||||||||
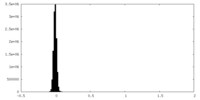
| Density Histograms |
-Half map: Half map of conformation 1 of the HCoV-NL63 RNA SL5 domain.
| File | emd_42813_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of conformation 1 of the HCoV-NL63 RNA SL5 domain. | ||||||||||||
| Projections & Slices |
| ||||||||||||
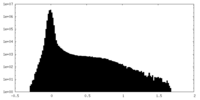
| Density Histograms |
- Sample components
Sample components
-Entire : HCoV-NL63 5' proximal stem-loop 5, conformation 1
| Entire | Name: HCoV-NL63 5' proximal stem-loop 5, conformation 1 |
|---|---|
| Components |
|
-Supramolecule #1: HCoV-NL63 5' proximal stem-loop 5, conformation 1
| Supramolecule | Name: HCoV-NL63 5' proximal stem-loop 5, conformation 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human coronavirus NL63 / Synthetically produced: Yes Human coronavirus NL63 / Synthetically produced: Yes |
| Molecular weight | Theoretical: 51.43 KDa |
-Macromolecule #1: HCoV-NL63 5' proximal stem-loop 5
| Macromolecule | Name: HCoV-NL63 5' proximal stem-loop 5 / type: rna / ID: 1 |
|---|---|
| Source (natural) | Organism:  Human coronavirus NL63 Human coronavirus NL63 |
| Sequence | String: GGUAAACUGG UUAGGCAAGU GUUGUAUUUU CUGUGUCUAA GCACUGGUGA UUCUGUUCAC UAGUGCAUAC AUUGAUAUUU AAGUGGUGUU CCGUCACUGC UUAUUGUGGA AGCAACGUUC UGUCGUUGUG GAAACCAAUA ACUGCUAACC AUGUUUUACC GENBANK: GENBANK: NC_005831.2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.28 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 8304 / Average exposure time: 3.61 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)