Yorodumi
Yorodumi+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | beta barrel / outer membrane protein / heme binding protein / heme transport / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationheme transport / siderophore transmembrane transport / heme transmembrane transporter activity / siderophore uptake transmembrane transporter activity / outer membrane / cell outer membrane / cellular response to iron ion Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Knejski P / Erramilli SK / Kossiakoff AA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Chaperone-assisted cryo-EM structure of P. aeruginosa PhuR reveals molecular basis for heme binding. Authors: Paweł P Knejski / Satchal K Erramilli / Anthony A Kossiakoff /   Abstract: Pathogenic bacteria, such as Pseudomonas aeruginosa, depend on scavenging heme for the acquisition of iron, an essential nutrient. The TonB-dependent transporter (TBDT) PhuR is the major heme uptake ...Pathogenic bacteria, such as Pseudomonas aeruginosa, depend on scavenging heme for the acquisition of iron, an essential nutrient. The TonB-dependent transporter (TBDT) PhuR is the major heme uptake protein in P. aeruginosa clinical isolates. However, a comprehensive understanding of heme recognition and TBDT transport mechanisms, especially PhuR, remains limited. In this study, we employed single-particle cryogenic electron microscopy (cryo-EM) and a phage display-generated synthetic antibody (sAB) as a fiducial marker to enable the determination of a high-resolution (2.5 Å) structure of PhuR with a bound heme. Notably, the structure reveals iron coordination by Y529 on a conserved extracellular loop, shedding light on the role of tyrosine in heme binding. Biochemical assays and negative-stain EM demonstrated that the sAB specifically targets the heme-bound state of PhuR. These findings provide insights into PhuR's heme binding and offer a template for developing conformation-specific sABs against outer membrane proteins (OMPs) for structure-function investigations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41255.map.gz emd_41255.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41255-v30.xml emd-41255-v30.xml emd-41255.xml emd-41255.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41255.png emd_41255.png | 115.3 KB | ||
| Masks |  emd_41255_msk_1.map emd_41255_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41255.cif.gz emd-41255.cif.gz | 6.7 KB | ||
| Others |  emd_41255_half_map_1.map.gz emd_41255_half_map_1.map.gz emd_41255_half_map_2.map.gz emd_41255_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41255 http://ftp.pdbj.org/pub/emdb/structures/EMD-41255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41255 | HTTPS FTP |
-Related structure data
| Related structure data |  8theMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41255.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41255.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.068 Å | ||||||||||||||||||||||||||||||||||||
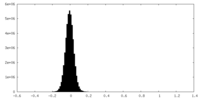
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41255_msk_1.map emd_41255_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
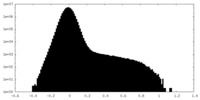
| Density Histograms |
-Half map: #2
| File | emd_41255_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41255_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heme bound P. aeruginosa PhuR with bound synthetic antibody
| Entire | Name: Heme bound P. aeruginosa PhuR with bound synthetic antibody |
|---|---|
| Components |
|
-Supramolecule #1: Heme bound P. aeruginosa PhuR with bound synthetic antibody
| Supramolecule | Name: Heme bound P. aeruginosa PhuR with bound synthetic antibody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Heme/hemoglobin uptake outer membrane receptor PhuR
| Macromolecule | Name: Heme/hemoglobin uptake outer membrane receptor PhuR / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.37032 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMGHHHHHH GENLYFQGGN AVPLTPTTIT ATRTEQAVDS VPSTVSVQTR EQLDRQNVNN IKELVRYEP GVSVGGAGQR AGITGYNIRG IDGNRILTQI DGVELPNDFF SGPYAQTHRN YVDPDIVKRV EILRGPASAL Y GSNAIGGA ...String: MKYLLPTAAA GLLLLAAQPA MAMGHHHHHH GENLYFQGGN AVPLTPTTIT ATRTEQAVDS VPSTVSVQTR EQLDRQNVNN IKELVRYEP GVSVGGAGQR AGITGYNIRG IDGNRILTQI DGVELPNDFF SGPYAQTHRN YVDPDIVKRV EILRGPASAL Y GSNAIGGA VSYFTLDPSD IIKDGKDVGA RLKAGYESAS HSWLTSATVA GRADDFDGLL HYGYRQGHET ESNGGHGGTG LS RSEANPE DADSYSLLGK LGWNYAEGSR FGLVFEKYKS DVDTDQKSAY GGPYDKGKPA IPPSMLPGGM YQWRKGNDTL TRE RYGLEH HFLLDSQVAD RIQWSLNYQL AKTDQATREF YYPITRKVLR TRDTTYKERL WVFDSQLDKS FAIGETEHLL SYGI NLKHQ KVTGMRSGTG TNLDTGADSP RDALERSSDF PDPTVKTYAL FAQDSISWND WTFTPGLRYD YTRMEPHITD EFLRT MKQS QNTAVDESDK KWHRVSPKFG VTYDFAQHYT WYGQYAQGFR TPTAKALYGR FENLQAGYHI EPNPNLKPEK SQSFET GLR GKFDEGSFGV AVFYNKYRDF IDEDALNTDS TGGNGQTFQS NNIERAVIKG VELKGRLELG AFGAPQGLYT QGSVAYA YG RNKDNGEPIN SVNPLTGVFG LGYDEADGNY GGLLSWTLVK RKDRVDDSTF HTPDGTASQF KTPGFGVLDL SAYYRLSK D LTLNAGLYNL TDKKYWLWDD VRGYDSVGEA SALAPANIDR LSQPGRNFAV NLVWDI UniProtKB: Heme/hemoglobin uptake outer membrane receptor PhuR |
-Macromolecule #2: Synthetic Antibody Heavy Chain
| Macromolecule | Name: Synthetic Antibody Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.525445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFYYYSIHWV RQAPGKGLEW VASISPYYGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSMNWYYSS PYGMSQGMDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFYYYSIHWV RQAPGKGLEW VASISPYYGS TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSMNWYYSS PYGMSQGMDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSCDKTHT |
-Macromolecule #3: Synthetic Antibody Light Chain
| Macromolecule | Name: Synthetic Antibody Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.212715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQGSSSPLTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQGSSSPLTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 4 / Number of copies: 1 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #5: (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[...
| Macromolecule | Name: (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-oxidanyl-5- ...Name: (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{S})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-4,5-bis(oxidanyl)oxane-2-carboxylic acid type: ligand / ID: 5 / Number of copies: 1 / Formula: CTQ |
|---|---|
| Molecular weight | Theoretical: 2.018542 KDa |
| Chemical component information |  ChemComp-CTQ: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 81 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 5031 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3) / Number images used: 292512 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3) |
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8the: |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)