+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Graphene sandwiched spike | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | C3 symmetry / SARS-CoV-2 / VIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Liu N / Xu J / Wang HW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Graphene sandwich-based biological specimen preparation for cryo-EM analysis. Authors: Jie Xu / Xiaoyin Gao / Liming Zheng / Xia Jia / Kui Xu / Yuwei Ma / Xiaoding Wei / Nan Liu / Hailin Peng / Hong-Wei Wang /  Abstract: High-quality specimen preparation plays a crucial role in cryo-electron microscopy (cryo-EM) structural analysis. In this study, we have developed a reliable and convenient technique called the ...High-quality specimen preparation plays a crucial role in cryo-electron microscopy (cryo-EM) structural analysis. In this study, we have developed a reliable and convenient technique called the graphene sandwich method for preparing cryo-EM specimens. This method involves using two layers of graphene films that enclose macromolecules on both sides, allowing for an appropriate ice thickness for cryo-EM analysis. The graphene sandwich helps to mitigate beam-induced charging effect and reduce particle motion compared to specimens prepared using the traditional method with graphene support on only one side, therefore improving the cryo-EM data quality. These advancements may open new opportunities to expand the use of graphene in the field of biological electron microscopy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38213.map.gz emd_38213.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38213-v30.xml emd-38213-v30.xml emd-38213.xml emd-38213.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38213.png emd_38213.png | 87.5 KB | ||
| Masks |  emd_38213_msk_1.map emd_38213_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38213.cif.gz emd-38213.cif.gz | 3.8 KB | ||
| Others |  emd_38213_half_map_1.map.gz emd_38213_half_map_1.map.gz emd_38213_half_map_2.map.gz emd_38213_half_map_2.map.gz | 37.5 MB 37.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38213 http://ftp.pdbj.org/pub/emdb/structures/EMD-38213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38213 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38213.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38213.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||
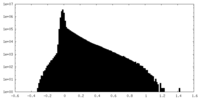
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38213_msk_1.map emd_38213_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
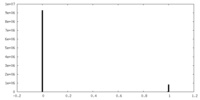
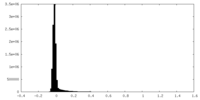
| Density Histograms |
-Half map: #1
| File | emd_38213_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
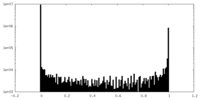
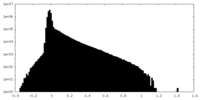
| Density Histograms |
-Half map: #2
| File | emd_38213_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
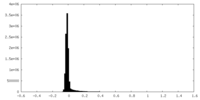
| Density Histograms |
- Sample components
Sample components
-Entire : spike of SARS-CoV-2
| Entire | Name: spike of SARS-CoV-2 |
|---|---|
| Components |
|
-Supramolecule #1: spike of SARS-CoV-2
| Supramolecule | Name: spike of SARS-CoV-2 / type: complex / ID: 1 / Parent: 0 / Details: graphene sandwiched |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 418743 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)