[English] 日本語
 Yorodumi
Yorodumi- EMDB-32954: Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of non gastric H,K-ATPase alpha2 K794A in (K+)E2-AlF state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationBasigin interactions / H+/K+-exchanging ATPase / Ion transport by P-type ATPases / Na+/K+-exchanging ATPase / P-type potassium:proton transporter activity / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / metal ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex ...Basigin interactions / H+/K+-exchanging ATPase / Ion transport by P-type ATPases / Na+/K+-exchanging ATPase / P-type potassium:proton transporter activity / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / metal ion transport / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / membrane repolarization / regulation of pH / sodium ion export across plasma membrane / intracellular potassium ion homeostasis / regulation of calcium ion transmembrane transport / potassium ion homeostasis / intracellular sodium ion homeostasis / regulation of cardiac muscle contraction by calcium ion signaling / response to metal ion / relaxation of cardiac muscle / Ion homeostasis / sodium ion transport / organelle membrane / potassium ion import across plasma membrane / monoatomic cation transmembrane transport / ATPase activator activity / blastocyst development / intercalated disc / lateral plasma membrane / sperm flagellum / sodium ion transmembrane transport / cardiac muscle contraction / ATP metabolic process / T-tubule / proton transmembrane transport / protein localization to plasma membrane / potassium ion transport / caveola / response to organic cyclic compound / sarcolemma / intracellular calcium ion homeostasis / ATPase binding / protein-macromolecule adaptor activity / regulation of gene expression / basolateral plasma membrane / response to hypoxia / cell adhesion / protein stabilization / protein heterodimerization activity / apical plasma membrane / innate immune response / protein kinase binding / ATP hydrolysis activity / ATP binding / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
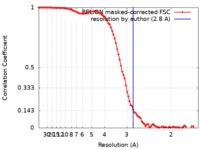
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Nakanishi H / Abe K | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure and function of H/K pump mutants reveal Na/K pump mechanisms. Authors: Victoria C Young / Hanayo Nakanishi / Dylan J Meyer / Tomohiro Nishizawa / Atsunori Oshima / Pablo Artigas / Kazuhiro Abe /   Abstract: Ion-transport mechanisms evolve by changing ion-selectivity, such as switching from Na to H selectivity in secondary-active transporters or P-type-ATPases. Here we study primary-active transport via ...Ion-transport mechanisms evolve by changing ion-selectivity, such as switching from Na to H selectivity in secondary-active transporters or P-type-ATPases. Here we study primary-active transport via P-type ATPases using functional and structural analyses to demonstrate that four simultaneous residue substitutions transform the non-gastric H/K pump, a strict H-dependent electroneutral P-type ATPase, into a bona fide Na-dependent electrogenic Na/K pump. Conversion of a H-dependent primary-active transporter into a Na-dependent one provides a prototype for similar studies of ion-transport proteins. Moreover, we solve the structures of the wild-type non-gastric H/K pump, a suitable drug target to treat cystic fibrosis, and of its Na/K pump-mimicking mutant in two major conformations, providing insight on how Na binding drives a concerted mechanism leading to Na/K pump phosphorylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32954.map.gz emd_32954.map.gz | 8.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32954-v30.xml emd-32954-v30.xml emd-32954.xml emd-32954.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32954_fsc.xml emd_32954_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32954.png emd_32954.png | 88.1 KB | ||
| Masks |  emd_32954_msk_1.map emd_32954_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_32954_half_map_1.map.gz emd_32954_half_map_1.map.gz emd_32954_half_map_2.map.gz emd_32954_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32954 http://ftp.pdbj.org/pub/emdb/structures/EMD-32954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32954 | HTTPS FTP |
-Validation report
| Summary document |  emd_32954_validation.pdf.gz emd_32954_validation.pdf.gz | 656.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32954_full_validation.pdf.gz emd_32954_full_validation.pdf.gz | 655.8 KB | Display | |
| Data in XML |  emd_32954_validation.xml.gz emd_32954_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_32954_validation.cif.gz emd_32954_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32954 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32954 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32954 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32954 | HTTPS FTP |
-Related structure data
| Related structure data |  7x21MC  7x20C  7x22C  7x23C  7x24C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32954.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32954.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32954_msk_1.map emd_32954_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32954_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32954_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : non gastric H,K-ATPase
+Supramolecule #1: non gastric H,K-ATPase
+Macromolecule #1: Potassium-transporting ATPase alpha chain 2
+Macromolecule #2: Sodium/potassium-transporting ATPase subunit beta-1
+Macromolecule #3: POTASSIUM ION
+Macromolecule #4: TETRAFLUOROALUMINATE ION
+Macromolecule #5: MAGNESIUM ION
+Macromolecule #6: CHOLESTEROL
+Macromolecule #7: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































 Homo sapiens (human)
Homo sapiens (human)