+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ACP1-KS-AT domains of mycobacterial Pks13 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mycolic acid synthesis / acyl carrier protein / ketosynthase / acyltransferase / multi-domain assembly / BIOSYNTHETIC PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information6-deoxyerythronolide-B synthase / erythronolide synthase activity / biosynthetic process / phosphopantetheine binding Similarity search - Function | ||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | ||||||||||||
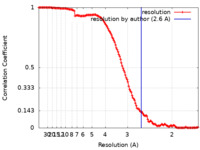
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | ||||||||||||
 Authors Authors | Kim SK / Dickinson MS / Finer-Moore JS / Rosenberg OS / Stroud RM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13. Authors: Sun Kyung Kim / Miles Sasha Dickinson / Janet Finer-Moore / Ziqiang Guan / Robyn M Kaake / Ignacia Echeverria / Jen Chen / Ernst H Pulido / Andrej Sali / Nevan J Krogan / Oren S Rosenberg / Robert M Stroud /  Abstract: The mycolic acid layer of the Mycobacterium tuberculosis cell wall is essential for viability and virulence, and the enzymes responsible for its synthesis are targets for antimycobacterial drug ...The mycolic acid layer of the Mycobacterium tuberculosis cell wall is essential for viability and virulence, and the enzymes responsible for its synthesis are targets for antimycobacterial drug development. Polyketide synthase 13 (Pks13) is a module encoding several enzymatic and transport functions that carries out the condensation of two different long-chain fatty acids to produce mycolic acids. We determined structures by cryogenic-electron microscopy of dimeric multi-enzyme Pks13 purified from mycobacteria under normal growth conditions, captured with native substrates. Structures define the ketosynthase (KS), linker and acyl transferase (AT) domains at 1.8 Å resolution and two alternative locations of the N-terminal acyl carrier protein. These structures suggest intermediate states on the pathway for substrate delivery to the KS domain. Other domains, visible at lower resolution, are flexible relative to the KS-AT core. The chemical structures of three bound endogenous long-chain fatty acid substrates were determined by electrospray ionization mass spectrometry. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27005.map.gz emd_27005.map.gz | 173.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27005-v30.xml emd-27005-v30.xml emd-27005.xml emd-27005.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27005_fsc.xml emd_27005_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_27005.png emd_27005.png | 61.9 KB | ||
| Others |  emd_27005_additional_1.map.gz emd_27005_additional_1.map.gz emd_27005_additional_2.map.gz emd_27005_additional_2.map.gz emd_27005_half_map_1.map.gz emd_27005_half_map_1.map.gz emd_27005_half_map_2.map.gz emd_27005_half_map_2.map.gz | 201.3 MB 186.1 MB 173.8 MB 173.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27005 http://ftp.pdbj.org/pub/emdb/structures/EMD-27005 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27005 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27005 | HTTPS FTP |
-Validation report
| Summary document |  emd_27005_validation.pdf.gz emd_27005_validation.pdf.gz | 930.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27005_full_validation.pdf.gz emd_27005_full_validation.pdf.gz | 930.5 KB | Display | |
| Data in XML |  emd_27005_validation.xml.gz emd_27005_validation.xml.gz | 21.2 KB | Display | |
| Data in CIF |  emd_27005_validation.cif.gz emd_27005_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27005 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27005 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27005 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27005 | HTTPS FTP |
-Related structure data
| Related structure data |  8cv1MC  7uk4C  8cuyC  8cuzC  8cv0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27005.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27005.map.gz / Format: CCP4 / Size: 219.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Phenix autosharpened map recommended contour at 5.3
| File | emd_27005_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix autosharpened map recommended contour at 5.3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
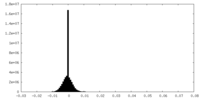
| Density Histograms |
-Supplemental map: emd 27005 additional 2.map
| File | emd_27005_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
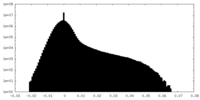
| Density Histograms |
-Half map: #2
| File | emd_27005_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
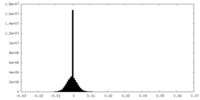
| Density Histograms |
-Half map: #1
| File | emd_27005_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
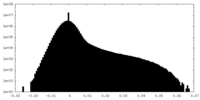
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterial polyketide synthase 13
| Entire | Name: Mycobacterial polyketide synthase 13 |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterial polyketide synthase 13
| Supramolecule | Name: Mycobacterial polyketide synthase 13 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: The gene for Mycobacterium smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP ...Details: The gene for Mycobacterium smegmatis polyketide synthase 13 (Pks13) was tagged with TEV-cleavable GFP at its C-terminus and purified from its natural source with anti-GFP nanobody beads. GFP was cleaved to yield the full-length Pks13. |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: Polyketide synthase PKS13
| Macromolecule | Name: Polyketide synthase PKS13 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: 6-deoxyerythronolide-B synthase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: ATCC 700084 / mc(2)155 Mycolicibacterium smegmatis MC2 155 (bacteria) / Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 194.668203 KDa |
| Sequence | String: MTVNEMREWL RNWVANATGQ SADAIDESTP MVELGLSSRD AVAMASDIED LTGVTLTATV AFRHPTIESL ATVIIEGEPE PEPYDEDED WSRTRDVEDI AIVGVATRFP GDLNTPDEMW EALLEGKDCV TDLPEDRWTE FLDEPRIAER VKKARTRGGY L TDIKGFDS ...String: MTVNEMREWL RNWVANATGQ SADAIDESTP MVELGLSSRD AVAMASDIED LTGVTLTATV AFRHPTIESL ATVIIEGEPE PEPYDEDED WSRTRDVEDI AIVGVATRFP GDLNTPDEMW EALLEGKDCV TDLPEDRWTE FLDEPRIAER VKKARTRGGY L TDIKGFDS EFFALSKMEA DNIDPQQRMA LELTWEALEH ARIPASSLRG ESVGVYIGSS TNDYSFLAMS DPSIAHPYAI TG TASSIIA NRVSYFYDFR GPSVAVDTAC SSSLVATHQG VQALRAGEAD VAIVGGVNAL VTPLVTVGFD EVGGVLAPDG RIK SFSSDA DGYARSEGGG MLVLKRISDA RRDGDQILAV IAGSAVNHDG RSNGLLAPNP DAQAEVLRKA YKDAGINPRD VDYI EAHGT GTILGDPIEA DALGRIVGKG RPADKPALLG AVKSNLGHLE SAAGAASLAK MTLALANDKL PPSINYAGPN PYIDF EKER LKVNDTVSDW PRYSGKAIAG VSGFGFGGAN AHVVMREVLA GDLVEPEPEP EPEAKPEKSE ADAVYVGGVR MDEYGE FID EDEPAEGGDA YPSYDEDSYE LPGITEAAQR LLEQAREELE AKEAEEPTKQ LVPLAVSAFL TSRKRQAAAE LADWIDS PE GRASSLESIG RSLSRRNHGR SRAVVLAHDH DEAIKGLRAL AEGKQHPSVL SADGPVTNGP VWVLAGFGAQ HRKMGKSL Y LRNEVFAEWI NKVDALIQDE RGYSILELIL DDNVDYTDAT CEYPIEVVQL VIFAIQIALG ELLRHHGAKP AAVVGQSLG EAAASYFAGG LSLADATRTI CSRSHLMGEG EAMLFGEYIR LMALVEYSAD EIKTVFSDYP DLEVCVYAAP TQTVIGGPPD QVDAIIARA ESEGKFARKF QTKGASHTQQ MDPLLGELAA ELQGIEPKPL TTGYFSTVHE GTFIRPGSAP IHDVDYWKKG L RHSVYFTQ GIRNAVDNGH TTFLELAPNP VALMQVGLTT ASAGLHDAQL IATLARKQDE VESMISAMAQ LYVHGHDLDF RT LFPRRSK GLAGALDFAN IPPTRFKRKE HWLPAHFTGD SSAVMPGNHV ATPDGRHVWE FVPRGKTDLA ALVKAAAAQV LPD AKLAAF EQRAVPADNA RLVTTLTRHP GGATVQVHAR VEESFTLVYD AIVARANGAG VTALPVAVGA GVAVSGDVAG EGAG ASVIE DDEPDAEILQ DNLTAGAGMG ADFQKWDPNS GETIGQRLGT IVGAAMGYEP EDLPWEVPLI ELGLDSLMAV RIKNR VEYD FDLPPIQLTA VRDANLYNVE ELIRYAIEHR DEVEQIAESQ KGKTAEEIAA EQSELLGGAS TVAELEAKLA EAGHPL AAK DSEDSENSED NAAGAAAAAE ASAVEGLEIP PPPTDPTGPG GAPIPPPPSD PSGPAQAASA TDAPAGTVNK ATAAAAA AK VLTQEAVTEA LGADVPPRDA AERVTFATWA IVTGKSPGGI FNELPTVSEE TAKKMAERLS ERAEGTITVE DVLGAKTI E GLATIVREQL EEGVVDGFVR TLRPPKEGSN AVPLFVFHPA GGSTVVYEPL MKRLPADVPV YGLERVEGSI EERAAEYVP KLLEMHKGPF VLAGWSLGGA LAYACAIGLK QSGADVRFVG LIDTVLPGEP IDQSKEGMRA RWDRYARFAE RTFNVEIPAI PYEELEKLD DEGQVKYVLE IVKESGVQIP GGIIEHQRTS YLDNRALDTV DIKPYDGHVT LYMADRYHDD AIVFEPAYAT R KPDGGWGS FVSDLEVVHI GGEHIQAIDE PYIAKVGAHM SEALNRIEAQ ASKEDGAK |
-Macromolecule #2: UNKNOWN LIGAND
| Macromolecule | Name: UNKNOWN LIGAND / type: ligand / ID: 2 / Number of copies: 2 / Formula: UNL |
|---|---|
| Molecular weight | Theoretical: 312.53 Da |
| Chemical component information | 
ChemComp-UNL: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 7 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV Details: Full length pks13 was concentrated to 1.5 mg mL-1 for cryo-EM grid preparation. 4.5 uL of sample was applied to freshly glow discharged holey carbon on gold R1.2/1.3 300 mesh Quantifoil ...Details: Full length pks13 was concentrated to 1.5 mg mL-1 for cryo-EM grid preparation. 4.5 uL of sample was applied to freshly glow discharged holey carbon on gold R1.2/1.3 300 mesh Quantifoil grids and blotted for 9 s with Whatman 1 filter paper at max humidity and 10oC in a FEI Mark IV Vitrobot, before vitrification in liquid nitrogen-cooled liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 7567 / Average exposure time: 5.9 sec. / Average electron dose: 45.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8cv1: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)