+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human KCC1 bound with VU0463271 In an outward-open state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC12A4 / VU0463271 / Outward-open state / TRANSPORT PROTEIN-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpotassium:chloride symporter activity / Cation-coupled Chloride cotransporters / chloride ion homeostasis / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane / monoatomic ion transport / potassium ion transmembrane transport / protein serine/threonine kinase binding / chloride transmembrane transport ...potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / chloride ion homeostasis / potassium ion homeostasis / cell volume homeostasis / potassium ion import across plasma membrane / monoatomic ion transport / potassium ion transmembrane transport / protein serine/threonine kinase binding / chloride transmembrane transport / chemical synaptic transmission / lysosomal membrane / synapse / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zhao YX / Cao EH | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structure of the human cation-chloride cotransport KCC1 in an outward-open state. Authors: Yongxiang Zhao / Jiemin Shen / Qinzhe Wang / Manuel Jose Ruiz Munevar / Pietro Vidossich / Marco De Vivo / Ming Zhou / Erhu Cao /   Abstract: Cation-chloride cotransporters (CCCs) catalyze electroneutral symport of Cl with Na and/or K across membranes. CCCs are fundamental in cell volume homeostasis, transepithelia ion movement, ...Cation-chloride cotransporters (CCCs) catalyze electroneutral symport of Cl with Na and/or K across membranes. CCCs are fundamental in cell volume homeostasis, transepithelia ion movement, maintenance of intracellular Cl concentration, and neuronal excitability. Here, we present a cryoelectron microscopy structure of human K-Cl cotransporter (KCC)1 bound with the VU0463271 inhibitor in an outward-open state. In contrast to many other amino acid-polyamine-organocation transporter cousins, our first outward-open CCC structure reveals that opening the KCC1 extracellular ion permeation path does not involve hinge-bending motions of the transmembrane (TM) 1 and TM6 half-helices. Instead, rocking of TM3 and TM8, together with displacements of TM4, TM9, and a conserved intracellular loop 1 helix, underlie alternate opening and closing of extracellular and cytoplasmic vestibules. We show that KCC1 intriguingly exists in one of two distinct dimeric states via different intersubunit interfaces. Our studies provide a blueprint for understanding the mechanisms of CCCs and their inhibition by small molecule compounds. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26116.map.gz emd_26116.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26116-v30.xml emd-26116-v30.xml emd-26116.xml emd-26116.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26116.png emd_26116.png | 30.5 KB | ||
| Filedesc metadata |  emd-26116.cif.gz emd-26116.cif.gz | 6.5 KB | ||
| Others |  emd_26116_additional_1.map.gz emd_26116_additional_1.map.gz | 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26116 http://ftp.pdbj.org/pub/emdb/structures/EMD-26116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26116 | HTTPS FTP |
-Related structure data
| Related structure data |  7ttiMC  7tthC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11047 (Title: Cryo-EM structure of human KCC1 bound with inhibitor VU0463271 EMPIAR-11047 (Title: Cryo-EM structure of human KCC1 bound with inhibitor VU0463271Data size: 3.9 TB Data #1: HumanKCC1-VU0463271 complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.035 Å | ||||||||||||||||||||||||||||||||||||
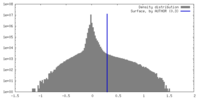
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26116_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
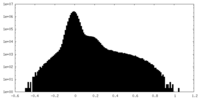
| Density Histograms |
- Sample components
Sample components
-Entire : human potassium-chloride cotransporter 1 bound with VU0463271
| Entire | Name: human potassium-chloride cotransporter 1 bound with VU0463271 |
|---|---|
| Components |
|
-Supramolecule #1: human potassium-chloride cotransporter 1 bound with VU0463271
| Supramolecule | Name: human potassium-chloride cotransporter 1 bound with VU0463271 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 4
| Macromolecule | Name: Solute carrier family 12 member 4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120.770492 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPHFTVVPVD GPRRGDYDNL EGLSWVDYGE RAELDDSDGH GNHRESSPFL SPLEASRGID YYDRNLALFE EELDIRPKVS SLLGKLVSY TNLTQGAKEH EEAESGEGTR RRAAEAPSMG TLMGVYLPCL QNIFGVILFL RLTWMVGTAG VLQALLIVLI C CCCTLLTA ...String: MPHFTVVPVD GPRRGDYDNL EGLSWVDYGE RAELDDSDGH GNHRESSPFL SPLEASRGID YYDRNLALFE EELDIRPKVS SLLGKLVSY TNLTQGAKEH EEAESGEGTR RRAAEAPSMG TLMGVYLPCL QNIFGVILFL RLTWMVGTAG VLQALLIVLI C CCCTLLTA ISMSAIATNG VVPAGGSYFM ISRSLGPEFG GAVGLCFYLG TTFAAAMYIL GAIEILLTYI APPAAIFYPS GA HDTSNAT LNNMRVYGTI FLTFMTLVVF VGVKYVNKFA SLFLACVIIS ILSIYAGGIK SIFDPPVFPV CMLGNRTLSR DQF DICAKT AVVDNETVAT QLWSFFCHSP NLTTDSCDPY FMLNNVTEIP GIPGAAAGVL QENLWSAYLE KGDIVEKHGL PSAD APSLK ESLPLYVVAD IATSFTVLVG IFFPSVTGIM AGSNRSGDLR DAQKSIPVGT ILAIITTSLV YFSSVVLFGA CIEGV VLRD KYGDGVSRNL VVGTLAWPSP WVIVIGSFFS TCGAGLQSLT GAPRLLQAIA KDNIIPFLRV FGHGKVNGEP TWALLL TAL IAELGILIAS LDMVAPILSM FFLMCYLFVN LACAVQTLLR TPNWRPRFKY YHWALSFLGM SLCLALMFVS SWYYALV AM LIAGMIYKYI EYQGAEKEWG DGIRGLSLSA ARYALLRLEE GPPHTKNWRP QLLVLLKLDE DLHVKYPRLL TFASQLKA G KGLTIVGSVI QGSFLESYGE AQAAEQTIKN MMEIEKVKGF CQVVVASKVR EGLAHLIQSC GLGGMRHNSV VLGWPYGWR QSEDPRAWKT FIDTVRCTTA AHLALLVPKN IAFYPSNHER YLEGHIDVWW IVHDGGMLML LPFLLRQHKV WRKCRMRIFT VAQMDDNSI QMKKDLAVFL YHLRLEAEVE VVEMHNSDIS AYTYERTLMM EQRSQMLRQM RLTKTERERE AQLVKDRHSA L RLESLYSD EEDESAVGAD KIQMTWTRDK YMTETWDPSH APDNFRELVH IKPDQSNVRR MHTAVKLNEV IVTRSHDARL VL LNMPGPP RNSEGDENYM EFLEVLTEGL ERVLLVRGGG REVITIYS UniProtKB: Solute carrier family 12 member 4 |
-Macromolecule #3: N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin...
| Macromolecule | Name: N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide type: ligand / ID: 3 / Number of copies: 2 / Formula: JUX |
|---|---|
| Molecular weight | Theoretical: 382.502 Da |
| Chemical component information | 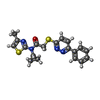 ChemComp-JUX: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)