[English] 日本語
 Yorodumi
Yorodumi- EMDB-24812: Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ION TRANSPORT / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / metal ion transmembrane transporter activity / cell body membrane / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity ...positive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / metal ion transmembrane transporter activity / cell body membrane / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / transepithelial chloride transport / potassium ion transmembrane transporter activity / intracellular chloride ion homeostasis / negative regulation of vascular wound healing / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / cell projection membrane / cellular response to chemokine / T cell chemotaxis / potassium ion homeostasis / intracellular sodium ion homeostasis / sodium ion import across plasma membrane / cellular response to potassium ion / cell volume homeostasis / hyperosmotic response / regulation of spontaneous synaptic transmission / gamma-aminobutyric acid signaling pathway / maintenance of blood-brain barrier / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / lateral plasma membrane / transport across blood-brain barrier / monoatomic ion transport / chloride transmembrane transport / basal plasma membrane / cytoplasmic vesicle membrane / sodium ion transmembrane transport / cell projection / cell periphery / Hsp90 protein binding / extracellular vesicle / protein-folding chaperone binding / cell body / basolateral plasma membrane / apical plasma membrane / neuron projection / neuronal cell body / protein kinase binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Zhao YX / Cao E | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide Authors: Zhao Y / Roy K / Vidossich P / Cancedda L / De Vivo M / Forbush B / Cao E | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24812.map.gz emd_24812.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24812-v30.xml emd-24812-v30.xml emd-24812.xml emd-24812.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24812.png emd_24812.png | 36.3 KB | ||
| Filedesc metadata |  emd-24812.cif.gz emd-24812.cif.gz | 6.3 KB | ||
| Others |  emd_24812_additional_1.map.gz emd_24812_additional_1.map.gz | 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24812 http://ftp.pdbj.org/pub/emdb/structures/EMD-24812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24812 | HTTPS FTP |
-Related structure data
| Related structure data |  7s1yMC  7s1xC  7s1zC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11048 (Title: Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide EMPIAR-11048 (Title: Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanideData size: 2.1 TB Data #1: Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24812.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24812.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
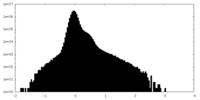
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_24812_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
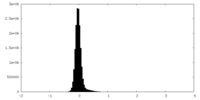
| Density Histograms |
- Sample components
Sample components
-Entire : hNKCC1 K289NA492E bound with bumetanide
| Entire | Name: hNKCC1 K289NA492E bound with bumetanide |
|---|---|
| Components |
|
-Supramolecule #1: hNKCC1 K289NA492E bound with bumetanide
| Supramolecule | Name: hNKCC1 K289NA492E bound with bumetanide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 2
| Macromolecule | Name: Solute carrier family 12 member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 131.841594 KDa |
| Recombinant expression | Organism: Mammalian expression vector BsrGI-MCS-pcDNA3.1 (others) |
| Sequence | String: GAMGSEPRPT APSSGAPGLA GVGETPSAAA LAAARVELPG TAVPSVPEDA APASRDGGGV RDEGPAAAGD GLGRPLGPTP SQSRFQVDL VSENAGRAAA AAAAAAAAAA AAGAGAGAKQ TPADGEASGE SEPAKGSEEA KGRFRVNFVD PAASSSAEDS L SDAAGVGV ...String: GAMGSEPRPT APSSGAPGLA GVGETPSAAA LAAARVELPG TAVPSVPEDA APASRDGGGV RDEGPAAAGD GLGRPLGPTP SQSRFQVDL VSENAGRAAA AAAAAAAAAA AAGAGAGAKQ TPADGEASGE SEPAKGSEEA KGRFRVNFVD PAASSSAEDS L SDAAGVGV DGPNVSFQNG GDTVLSEGSS LHSGGGGSGH HQHYYYDTHT NTYYLRTFGH NTMDAVPRID HYRHTAAQLG EK LLRPSLA ELHDELEKEP FEDGFANGEE STPTRDAVVT YTAESKGVVK FGWINGVLVR CMLNIWGVML FIRLSWIVGQ AGI GLSVLV IMMATVVTTI TGLSTSAIAT NGFVRGGGAY YLISRSLGPE FGGAIGLIFA FANAVAVAMY VVGFAETVVE LLKE HSILM IDEINDIRII GAITVVILLG ISVAGMEWEA KAQIVLLVIL LLAIGDFVIG TFIPLESKKP KGFFGYKSEI FNENF GPDF REEETFFSVF EIFFPAATGI LAGANISGDL ADPQSALPKG TLLAILITTL VYVGIAVSVG SCVVRDATGN VNDTIV TEL TNCTSAACKL NFDFSSCESS PCSYGLMNNF QVMSMVSGFT PLISAGIFSA TLSSALASLV SAPKIFQALC KDNIYPA FQ MFAKGYGKNN EPLRGYILTF LIALGFILIA ELNVIAPIIS NFFLASYALI NFSVFHASLA KSPGWRPAFK YYNMWISL L GAILCCIVMF VINWWAALLT YVIVLGLYIY VTYKKPDVNW GSSTQALTYL NALQHSIRLS GVEDHVKNFR PQCLVMTGA PNSRPALLHL VHDFTKNVGL MICGHVHMGP RRQAMKEMSI DQAKYQRWLI KNKMKAFYAP VHADDLREGA QYLMQAAGLG RMKPNTLVL GFKKDWLQAD MRDVDMYINL FHDAFDIQYG VVVIRLKEGL DISHLQGQEE LLSSQEKSPG TKDVVVSVEY S KKSDLDTS KPLSEKPITH KVEEEDGKTA TQPLLKKESK GPIVPLNVAD QKLLEASTQF QKKQGKNTID VWWLFDDGGL TL LIPYLLT TKKKWKDCKI RVFIGGKINR IDHDRRAMAT LLSKFRIDFS DIMVLGDINT KPKKENIIAF EEIIEPYRLH EDD KEQDIA DKMKEDEPWR ITDNELELYK TKTYRQIRLN ELLKEHSSTA NIIVMSLPVA RKGAVSSALY MAWLEALSKD LPPI LLVRG NHQSVLTFYS UniProtKB: Solute carrier family 12 member 2 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid
| Macromolecule | Name: 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid / type: ligand / ID: 4 / Number of copies: 2 / Formula: 82U |
|---|---|
| Molecular weight | Theoretical: 364.416 Da |
| Chemical component information | 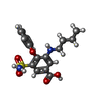 ChemComp-82U: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.175 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7s1y: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)