+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Human NKCC1 K289NA492EL671C | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / cell body membrane / metal ion transmembrane transporter activity / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity ...positive regulation of cell volume / positive regulation of aspartate secretion / transepithelial ammonium transport / regulation of matrix metallopeptidase secretion / cell body membrane / metal ion transmembrane transporter activity / inorganic anion import across plasma membrane / inorganic cation import across plasma membrane / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / potassium ion transmembrane transporter activity / transepithelial chloride transport / intracellular chloride ion homeostasis / ammonium transmembrane transport / negative regulation of vascular wound healing / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / cell projection membrane / cellular response to chemokine / T cell chemotaxis / potassium ion homeostasis / intracellular sodium ion homeostasis / sodium ion import across plasma membrane / cell volume homeostasis / cellular response to potassium ion / hyperosmotic response / regulation of spontaneous synaptic transmission / maintenance of blood-brain barrier / gamma-aminobutyric acid signaling pathway / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / lateral plasma membrane / transport across blood-brain barrier / monoatomic ion transport / sodium ion transmembrane transport / basal plasma membrane / chloride transmembrane transport / cytoplasmic vesicle membrane / cell periphery / cell projection / Hsp90 protein binding / extracellular vesicle / protein-folding chaperone binding / cell body / basolateral plasma membrane / neuron projection / apical plasma membrane / neuronal cell body / protein kinase binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhao YX / Cao EH | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide Authors: Zhao Y / Roy K / Vidossich P / Cancedda L / De Vivo M / Forbush B / Cao E | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24813.map.gz emd_24813.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24813-v30.xml emd-24813-v30.xml emd-24813.xml emd-24813.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24813.png emd_24813.png | 39.4 KB | ||
| Filedesc metadata |  emd-24813.cif.gz emd-24813.cif.gz | 6.1 KB | ||
| Others |  emd_24813_additional_1.map.gz emd_24813_additional_1.map.gz | 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24813 http://ftp.pdbj.org/pub/emdb/structures/EMD-24813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24813 | HTTPS FTP |
-Validation report
| Summary document |  emd_24813_validation.pdf.gz emd_24813_validation.pdf.gz | 483.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24813_full_validation.pdf.gz emd_24813_full_validation.pdf.gz | 482.3 KB | Display | |
| Data in XML |  emd_24813_validation.xml.gz emd_24813_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_24813_validation.cif.gz emd_24813_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24813 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24813 | HTTPS FTP |
-Related structure data
| Related structure data |  7s1zMC  7s1xC  7s1yC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24813.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24813.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
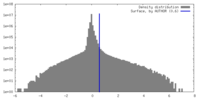
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_24813_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
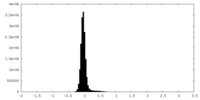
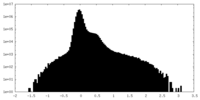
| Density Histograms |
- Sample components
Sample components
-Entire : Human NKCC1 K289NA492EL671C
| Entire | Name: Human NKCC1 K289NA492EL671C |
|---|---|
| Components |
|
-Supramolecule #1: Human NKCC1 K289NA492EL671C
| Supramolecule | Name: Human NKCC1 K289NA492EL671C / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 2
| Macromolecule | Name: Solute carrier family 12 member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 131.888641 KDa |
| Recombinant expression | Organism: Mammalian expression vector BsrGI-MCS-pcDNA3.1 (others) |
| Sequence | String: GAMGSEPRPT APSSGAPGLA GVGETPSAAA LAAARVELPG TAVPSVPEDA APASRDGGGV RDEGPAAAGD GLGRPLGPTP SQSRFQVDL VSENAGRAAA AAAAAAAAAA AAGAGAGAKQ TPADGEASGE SEPAKGSEEA KGRFRVNFVD PAASSSAEDS L SDAAGVGV ...String: GAMGSEPRPT APSSGAPGLA GVGETPSAAA LAAARVELPG TAVPSVPEDA APASRDGGGV RDEGPAAAGD GLGRPLGPTP SQSRFQVDL VSENAGRAAA AAAAAAAAAA AAGAGAGAKQ TPADGEASGE SEPAKGSEEA KGRFRVNFVD PAASSSAEDS L SDAAGVGV DGPNVSFQNG GDTVLSEGSS LHSGGGGGSG HHQHYYYDTH TNTYYLRTFG HNTMDAVPRI DHYRHTAAQL GE KLLRPSL AELHDELEKE PFEDGFANGE ESTPTRDAVV TYTAESKGVV KFGWINGVLV RCMLNIWGVM LFIRLSWIVG QAG IGLSVL VIMMATVVTT ITGLSTSAIA TNGFVRGGGA YYLISRSLGP EFGGAIGLIF AFANAVAVAM YVVGFAETVV ELLK EHSIL MIDEINDIRI IGAITVVILL GISVAGMEWE AKAQIVLLVI LLLAIGDFVI GTFIPLESKK PKGFFGYKSE IFNEN FGPD FREEETFFSV FEIFFPAATG ILAGANISGD LADPQSALPK GTLLAILITT LVYVGIAVSV GSCVVRDATG NVNDTI VTE LTNCTSAACK LNFDFSSCES SPCSYGLMNN FQVMSMVSGF TPLISAGIFS ATLSSALASL VSAPKIFQAL CKDNIYP AF QMFAKGYGKN NEPLRGYILT FLIALGFILI AECNVIAPII SNFFLASYAL INFSVFHASL AKSPGWRPAF KYYNMWIS L LGAILCCIVM FVINWWAALL TYVIVLGLYI YVTYKKPDVN WGSSTQALTY LNALQHSIRL SGVEDHVKNF RPQCLVMTG APNSRPALLH LVHDFTKNVG LMICGHVHMG PRRQAMKEMS IDQAKYQRWL IKNKMKAFYA PVHADDLREG AQYLMQAAGL GRMKPNTLV LGFKKDWLQA DMRDVDMYIN LFHDAFDIQY GVVVIRLKEG LDISHLQGQE ELLSSQEKSP GTKDVVVSVE Y SKKSDLDT SKPLSEKPIT HKVEEEDGKT ATQPLLKKES KGPIVPLNVA DQKLLEASTQ FQKKQGKNTI DVWWLFDDGG LT LLIPYLL TTKKKWKDCK IRVFIGGKIN RIDHDRRAMA TLLSKFRIDF SDIMVLGDIN TKPKKENIIA FEEIIEPYRL HED DKEQDI ADKMKEDEPW RITDNELELY KTKTYRQIRL NELLKEHSST ANIIVMSLPV ARKGAVSSAL YMAWLEALSK DLPP ILLVR GNHQSVLTFY S UniProtKB: Solute carrier family 12 member 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.175 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)