[English] 日本語
 Yorodumi
Yorodumi- EMDB-24688: Structures of Tweety Homolog Proteins TTYH2 and TTYH3 in lipid na... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24688 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structures of Tweety Homolog Proteins TTYH2 and TTYH3 in lipid nanodiscs | |||||||||
 Map data Map data | Tweety Homolog Proteins TTYH2 and TTYH3 in lipid nanodiscs | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TTYH2 / Cisdimer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvolume-sensitive chloride channel activity / L-glutamate transmembrane transport / Stimuli-sensing channels / chloride channel complex / calcium ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Li B / Brohawn SG | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca-dependent switch from intra- to intermembrane dimerization. Authors: Baobin Li / Christopher M Hoel / Stephen G Brohawn /  Abstract: Tweety homologs (TTYHs) comprise a conserved family of transmembrane proteins found in eukaryotes with three members (TTYH1-3) in vertebrates. They are widely expressed in mammals including at high ...Tweety homologs (TTYHs) comprise a conserved family of transmembrane proteins found in eukaryotes with three members (TTYH1-3) in vertebrates. They are widely expressed in mammals including at high levels in the nervous system and have been implicated in cancers and other diseases including epilepsy, chronic pain, and viral infections. TTYHs have been reported to form Ca- and cell volume-regulated anion channels structurally distinct from any characterized protein family with potential roles in cell adhesion, migration, and developmental signaling. To provide insight into TTYH family structure and function, we determined cryo-EM structures of Mus musculus TTYH2 and TTYH3 in lipid nanodiscs. TTYH2 and TTYH3 adopt a previously unobserved fold which includes an extended extracellular domain with a partially solvent exposed pocket that may be an interaction site for hydrophobic molecules. In the presence of Ca, TTYH2 and TTYH3 form homomeric cis-dimers bridged by extracellularly coordinated Ca. Strikingly, in the absence of Ca, TTYH2 forms trans-dimers that span opposing membranes across a ~130 Å intermembrane space as well as a monomeric state. All TTYH structures lack ion conducting pathways and we do not observe TTYH2-dependent channel activity in cells. We conclude TTYHs are not pore forming subunits of anion channels and their function may involve Ca-dependent changes in quaternary structure, interactions with hydrophobic molecules near the extracellular membrane surface, and/or association with additional protein partners. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24688.map.gz emd_24688.map.gz | 57.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24688-v30.xml emd-24688-v30.xml emd-24688.xml emd-24688.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24688.png emd_24688.png | 47.8 KB | ||
| Filedesc metadata |  emd-24688.cif.gz emd-24688.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24688 http://ftp.pdbj.org/pub/emdb/structures/EMD-24688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24688 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24688 | HTTPS FTP |
-Related structure data
| Related structure data |  7rttMC  7rtuC  7rtvC  7rtwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10842 (Title: Cryo-EM structure of a TTYH2 cis-dimer / Data size: 3.4 TB EMPIAR-10842 (Title: Cryo-EM structure of a TTYH2 cis-dimer / Data size: 3.4 TBData #1: Unaligned multi-frame micrographs of ttyh2 in cis-dimer [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24688.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24688.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tweety Homolog Proteins TTYH2 and TTYH3 in lipid nanodiscs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.137 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
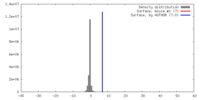
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TTYH2
| Entire | Name: TTYH2 |
|---|---|
| Components |
|
-Supramolecule #1: TTYH2
| Supramolecule | Name: TTYH2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein tweety homolog 2
| Macromolecule | Name: Protein tweety homolog 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.941434 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: PAARVEYIAP WWVVWLHSVP HLGLRLQRVD STFSPGDETY QESLLFLGVL AAIGLGLNLI FLTVYLVCTC CCRRDHTVQT KQQESCCVT WTAVVAGLLC CAAVGVGFYG NSETNDGMHQ LIYSLDNANH TFSGMDELVS ANTQRMKVDL EQHLARLSEI I AARGDYIQ ...String: PAARVEYIAP WWVVWLHSVP HLGLRLQRVD STFSPGDETY QESLLFLGVL AAIGLGLNLI FLTVYLVCTC CCRRDHTVQT KQQESCCVT WTAVVAGLLC CAAVGVGFYG NSETNDGMHQ LIYSLDNANH TFSGMDELVS ANTQRMKVDL EQHLARLSEI I AARGDYIQ TLKFMQQMAG NVVSQLSGLP VWREVTTQLT KLSHQTAYVE YYRWLSYLLL FILDLVICLV TCLGLARRSK CL LASMLCC GILTLILSWA SLAADAAAAV GTSDFCMAPD IYILNNTGSQ INSEVTRYYL HCSQSLISPF QQSLTTFQRS LTT MQIQVG GLLQFAVPLF PTAEKDLLGI QLLLNNSEIS LHQLTAMLDC RGLHKDYLDA LTGICYDGIE GLLFLGLFSL LAAL AFSTL TCAGPRAWKY FINRDRDYDD IDDDDPFNPQ ARRIAAHNPT RGQLHSFCSY SSGLGSQCSL QPPSQTISNA PVSEY MNQA ILFGGNPRYE NVPLIGRGSP PPTYSPSMRP TYMSVADEHL RHYEFPSSNS LEVLFQ UniProtKB: Protein tweety homolog 2 |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM CaCl2 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.0038 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7rtt: |
 Movie
Movie Controller
Controller












 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)