+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23996 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of mGlu2 - Gi Complex | |||||||||
 Map data Map data | Composite CryoEM Map of mGlu2 - Heterotrimeric Gi Complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Metabotropic Glutamate Receptor 2 (mGlu2) (mGluR2) / Family C G protein-coupled receptor (GPCR) / Heterotrimeric G protein / CryoEM structure / MEMBRANE PROTEIN / MEMBRANE PROTEIN-SIGNALING PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of response to drug / group II metabotropic glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate secretion / long-term synaptic depression / glutamate receptor activity ...regulation of response to drug / group II metabotropic glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / glutamate secretion / long-term synaptic depression / glutamate receptor activity / regulation of glutamate secretion / astrocyte projection / cellular response to stress / regulation of dopamine secretion / regulation of synaptic transmission, glutamatergic / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / cellular response to forskolin / regulation of mitotic spindle organization / response to cocaine / calcium channel regulator activity / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor activity / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / presynaptic membrane / Ca2+ pathway / fibroblast proliferation / scaffold protein binding / gene expression / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Ras protein signal transduction / postsynaptic membrane / Extra-nuclear estrogen signaling / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / cell population proliferation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Seven AB / Barros-Alvarez X | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: G-protein activation by a metabotropic glutamate receptor. Authors: Alpay B Seven / Ximena Barros-Álvarez / Marine de Lapeyrière / Makaía M Papasergi-Scott / Michael J Robertson / Chensong Zhang / Robert M Nwokonko / Yang Gao / Justin G Meyerowitz / Jean- ...Authors: Alpay B Seven / Ximena Barros-Álvarez / Marine de Lapeyrière / Makaía M Papasergi-Scott / Michael J Robertson / Chensong Zhang / Robert M Nwokonko / Yang Gao / Justin G Meyerowitz / Jean-Philippe Rocher / Dominik Schelshorn / Brian K Kobilka / Jesper M Mathiesen / Georgios Skiniotis /    Abstract: Family C G-protein-coupled receptors (GPCRs) operate as obligate dimers with extracellular domains that recognize small ligands, leading to G-protein activation on the transmembrane (TM) domains of ...Family C G-protein-coupled receptors (GPCRs) operate as obligate dimers with extracellular domains that recognize small ligands, leading to G-protein activation on the transmembrane (TM) domains of these receptors by an unknown mechanism. Here we show structures of homodimers of the family C metabotropic glutamate receptor 2 (mGlu2) in distinct functional states and in complex with heterotrimeric G. Upon activation of the extracellular domain, the two transmembrane domains undergo extensive rearrangement in relative orientation to establish an asymmetric TM6-TM6 interface that promotes conformational changes in the cytoplasmic domain of one protomer. Nucleotide-bound G can be observed pre-coupled to inactive mGlu2, but its transition to the nucleotide-free form seems to depend on establishing the active-state TM6-TM6 interface. In contrast to family A and B GPCRs, G-protein coupling does not involve the cytoplasmic opening of TM6 but is facilitated through the coordination of intracellular loops 2 and 3, as well as a critical contribution from the C terminus of the receptor. The findings highlight the synergy of global and local conformational transitions to facilitate a new mode of G-protein activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23996.map.gz emd_23996.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23996-v30.xml emd-23996-v30.xml emd-23996.xml emd-23996.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23996.png emd_23996.png | 97.4 KB | ||
| Filedesc metadata |  emd-23996.cif.gz emd-23996.cif.gz | 7.1 KB | ||
| Others |  emd_23996_additional_1.map.gz emd_23996_additional_1.map.gz emd_23996_additional_2.map.gz emd_23996_additional_2.map.gz emd_23996_additional_3.map.gz emd_23996_additional_3.map.gz emd_23996_additional_4.map.gz emd_23996_additional_4.map.gz | 2.9 MB 1 MB 2.2 MB 2.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23996 http://ftp.pdbj.org/pub/emdb/structures/EMD-23996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23996 | HTTPS FTP |
-Related structure data
| Related structure data |  7mtsMC  7mtqC  7mtrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10979 (Title: CryoEM Structure of mGlu2 - Gi Complex / Data size: 21.6 TB EMPIAR-10979 (Title: CryoEM Structure of mGlu2 - Gi Complex / Data size: 21.6 TBData #1: Unaligned multi-frame micrographs of mGlu2 - Gi Complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23996.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23996.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite CryoEM Map of mGlu2 - Heterotrimeric Gi Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8677 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Local and focused refinement of mGlu2 VFT and CRD domains
| File | emd_23996_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local and focused refinement of mGlu2 VFT and CRD domains | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local and focused refinement of G-beta-gamma
| File | emd_23996_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local and focused refinement of G-beta-gamma | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local and focused refinement of mGlu2 CRD and 7TM domains
| File | emd_23996_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local and focused refinement of mGlu2 CRD and 7TM domains | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local and focused refinement of mGlu2 7TM domain and G-alpha-i
| File | emd_23996_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local and focused refinement of mGlu2 7TM domain and G-alpha-i | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mGlu2 - Heterotrimeric Gi complex
| Entire | Name: mGlu2 - Heterotrimeric Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: mGlu2 - Heterotrimeric Gi complex
| Supramolecule | Name: mGlu2 - Heterotrimeric Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Heterotrimeric G protein coupled metabotropic glutamate receptor 2 bound to Ago-PAM ADX55164 and L-Glutamate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 277 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 2
| Macromolecule | Name: Metabotropic glutamate receptor 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.932445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEGPAKKVLT LEGDLVLGGL FPVHQKGGPA EDCGPVNEHR GIQRLEAMLF ALDRINRDPH LLPGVRLGAH ILDSCSKDTH ALEQALDFV RASLSRGADG SRHICPDGSY ATHGDAPTAI TGVIGGSYSD VSIQVANLLR LFQIPQISYA STSAKLSDKS R YDYFARTV ...String: AEGPAKKVLT LEGDLVLGGL FPVHQKGGPA EDCGPVNEHR GIQRLEAMLF ALDRINRDPH LLPGVRLGAH ILDSCSKDTH ALEQALDFV RASLSRGADG SRHICPDGSY ATHGDAPTAI TGVIGGSYSD VSIQVANLLR LFQIPQISYA STSAKLSDKS R YDYFARTV PPDFFQAKAM AEILRFFNWT YVSTVASEGD YGETGIEAFE LEARARNICV ATSEKVGRAM SRAAFEGVVR AL LQKPSAR VAVLFTRSED ARELLAASQR LNASFTWVAS DGWGALESVV AGSEGAAEGA ITIELASYPI SDFASYFQSL DPW NNSRNP WFREFWEQRF RCSFRQRDCA AHSLRAVPFE QESKIMFVVN AVYAMAHALH NMHRALCPNT TRLCDAMRPV NGRR LYKDF VLNVKFDAPF RPADTHNEVR FDRFGDGIGR YNIFTYLRAG SGRYRYQKVG YWAEGLTLDT SLIPWASPSA GPLPA SRCS EPCLQNEVKS VQPGEVCCWL CIPCQPYEYR LDEFTCADCG LGYWPNASLT GCFELPQEYI RWGDAWAVGP VTIACL GAL ATLFVLGVFV RHNATPVVKA SGRELCYILL GGVFLCYCMT FIFIAKPSTA VCTLRRLGLG TAFSVCYSAL LTKTNRI AR IFGGAREGAQ RPRFISPASQ VAICLALISG QLLIVVAWLV VEAPGTGKET APERREVVTL RCNHRDASML GSLAYNVL L IALCTLYAFK TRKCPENFNE AKFIGFTMYT TCIIWLAFLP IFYVTSSDYR VQTTTMCVSV SLSGSVVLGC LFAPKLHII LFQPQKNVVS HRAPTSRFGS AAARASSSLG QGSGSQFVPT VCNGREVVDS TTSSL UniProtKB: Metabotropic glutamate receptor 2 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.342785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethy...
| Macromolecule | Name: 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine type: ligand / ID: 5 / Number of copies: 1 / Formula: ZQY |
|---|---|
| Molecular weight | Theoretical: 425.507 Da |
| Chemical component information | 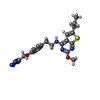 ChemComp-ZQY: |
-Macromolecule #6: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 6 / Number of copies: 2 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2) / Details: A composite map of four locally refined maps. / Number images used: 799323 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)