[English] 日本語
 Yorodumi
Yorodumi- EMDB-2289: Second 3D model of wild type MjHSP16.5 at room temperature by CryoEM. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2289 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Second 3D model of wild type MjHSP16.5 at room temperature by CryoEM. | |||||||||
 Map data Map data | Second model for reconstruction of wild type MjHSP16.5 at room temperature | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Small heat shock protein / HSP16.5 | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to stress / protein complex oligomerization / response to salt stress / protein folding chaperone / response to hydrogen peroxide / unfolded protein binding / protein folding / response to heat / protein stabilization / protein-containing complex ...response to stress / protein complex oligomerization / response to salt stress / protein folding chaperone / response to hydrogen peroxide / unfolded protein binding / protein folding / response to heat / protein stabilization / protein-containing complex / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Methanocaldococcus jannaschii (archaea) Methanocaldococcus jannaschii (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.0 Å | |||||||||
 Authors Authors | Roy A Q / Yan Z / Andrew L / Ian W / Ehmke P / Fei S | |||||||||
 Citation Citation |  Journal: Philos Trans R Soc Lond B Biol Sci / Year: 2013 Journal: Philos Trans R Soc Lond B Biol Sci / Year: 2013Title: Changes in the quaternary structure and function of MjHSP16.5 attributable to deletion of the IXI motif and introduction of the substitution, R107G, in the α-crystallin domain. Authors: Roy A Quinlan / Yan Zhang / Andrew Lansbury / Ian Williamson / Ehmke Pohl / Fei Sun /  Abstract: The archael small heat-shock protein (sHSP), MjHSP16.5, forms a 24-subunit oligomer with octahedral symmetry. Here, we demonstrate that the IXI motif present in the C-terminal domain is necessary for ...The archael small heat-shock protein (sHSP), MjHSP16.5, forms a 24-subunit oligomer with octahedral symmetry. Here, we demonstrate that the IXI motif present in the C-terminal domain is necessary for the oligomerization of MjHSP16.5. Removal increased the in vitro chaperone activity with citrate synthase as the client protein. Less predictable were the effects of the R107G substitution in MjHSP16.5 because of the differences in the oligomerization of metazoan and non-metazoan sHSPs. We present the crystal structure for MjHSP16.5 R107G and compare this with an improved (2.5 Å) crystal structure for wild-type (WT) MjHSP16.5. Although no significant structural differences were found in the crystal, using cryo-electron microscopy, we identified two 24mer species with octahedral symmetry for the WT MjHSP16.5 both at room temperature and at 60°C, all showing two major species with the same diameter of 12.4 nm. Similarly, at room temperature, there are also two kinds of 12.4 nm oligomers for R107G MjHSP16.5, but in the 60°C sample, a larger 24mer species with a diameter of 13.6 nm was observed with significant changes in the fourfold symmetry axis and dimer-dimer interface. This highly conserved arginine, therefore, contributes to the quaternary organization of non-metazoan sHSP oligomers. Potentially, the R107G substitution has functional consequences as R107G MjHSP16.5 was far superior to the WT protein in protecting βL-crystallin against heat-induced aggregation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2289.map.gz emd_2289.map.gz | 56.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2289-v30.xml emd-2289-v30.xml emd-2289.xml emd-2289.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  2289_EMDB-2289.tif 2289_EMDB-2289.tif | 118.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2289 http://ftp.pdbj.org/pub/emdb/structures/EMD-2289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2289 | HTTPS FTP |
-Related structure data
| Related structure data |  2288C 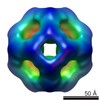 2290C  2291C  2292C  2293C  2294C  2295C  4i88C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2289.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2289.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second model for reconstruction of wild type MjHSP16.5 at room temperature | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.933 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
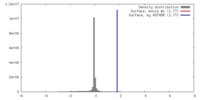
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : wild type small heat shock protein (sHSP) HSP16.5 from Methanocal...
| Entire | Name: wild type small heat shock protein (sHSP) HSP16.5 from Methanocaldococcus jannaschii (MjHSP16.5) |
|---|---|
| Components |
|
-Supramolecule #1000: wild type small heat shock protein (sHSP) HSP16.5 from Methanocal...
| Supramolecule | Name: wild type small heat shock protein (sHSP) HSP16.5 from Methanocaldococcus jannaschii (MjHSP16.5) type: sample / ID: 1000 / Oligomeric state: 24mer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 396 KDa / Theoretical: 396 KDa |
-Macromolecule #1: MjHSP16.5
| Macromolecule | Name: MjHSP16.5 / type: protein_or_peptide / ID: 1 / Name.synonym: small heat shock protein (sHSP) from HSP16.5 / Number of copies: 24 / Oligomeric state: 24mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Methanocaldococcus jannaschii (archaea) / Location in cell: cytoplasmic Methanocaldococcus jannaschii (archaea) / Location in cell: cytoplasmic |
| Molecular weight | Experimental: 16.5 KDa / Theoretical: 16.5 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Small heat shock protein HSP16.5 / GO: cytoplasm, response to stress InterPro: Alpha crystallin/Hsp20 domain, HSP20-like chaperone |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10mM HEPES, 100mM NaCl. |
| Grid | Details: GIG holey grids (LifeTrust, China) were treated with a glow discharge machine (Master Plasmer) |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 93 K / Instrument: FEI VITROBOT MARK IV Method: The samples were blotted for 2 s with blot force 2 at 100% humidity. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Average: 85 K |
| Date | Sep 28, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 1160 / Average electron dose: 20 e/Å2 Details: Electron micrograph exposures were made with the automatic collection package Leginon. Bits/pixel: 32 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder: liquid nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using an automatic selection program Gautomatch developed in Fei Sun lab (to be published). Octahedron symmetry were imposed during 3D reconstructing. |
|---|---|
| CTF correction | Details: Each image |
| Final reconstruction | Applied symmetry - Point group: O (octahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 12.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN1 Details: The final reconstructed density map was further sharpened by application of an amplitude correction algorithm in the program BFACTOR. Number images used: 4356 |
| Final two d classification | Number classes: 193 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)