+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22878 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average structure of E. coli polysome | |||||||||
 Map data Map data | E. coli polysome | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
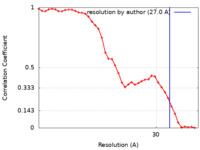
| Method | subtomogram averaging / cryo EM / Resolution: 27.0 Å | |||||||||
 Authors Authors | Chang YJ / Liu J / Xiang YJ / Jacobs-Wagner C | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Interconnecting solvent quality, transcription, and chromosome folding in Escherichia coli. Authors: Yingjie Xiang / Ivan V Surovtsev / Yunjie Chang / Sander K Govers / Bradley R Parry / Jun Liu / Christine Jacobs-Wagner /  Abstract: All cells fold their genomes, including bacterial cells, where the chromosome is compacted into a domain-organized meshwork called the nucleoid. How compaction and domain organization arise is not ...All cells fold their genomes, including bacterial cells, where the chromosome is compacted into a domain-organized meshwork called the nucleoid. How compaction and domain organization arise is not fully understood. Here, we describe a method to estimate the average mesh size of the nucleoid in Escherichia coli. Using nucleoid mesh size and DNA concentration estimates, we find that the cytoplasm behaves as a poor solvent for the chromosome when the cell is considered as a simple semidilute polymer solution. Monte Carlo simulations suggest that a poor solvent leads to chromosome compaction and DNA density heterogeneity (i.e., domain formation) at physiological DNA concentration. Fluorescence microscopy reveals that the heterogeneous DNA density negatively correlates with ribosome density within the nucleoid, consistent with cryoelectron tomography data. Drug experiments, together with past observations, suggest the hypothesis that RNAs contribute to the poor solvent effects, connecting chromosome compaction and domain formation to transcription and intracellular organization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22878.map.gz emd_22878.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22878-v30.xml emd-22878-v30.xml emd-22878.xml emd-22878.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22878_fsc.xml emd_22878_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22878.png emd_22878.png | 63.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22878 http://ftp.pdbj.org/pub/emdb/structures/EMD-22878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22878 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22878 | HTTPS FTP |
-Validation report
| Summary document |  emd_22878_validation.pdf.gz emd_22878_validation.pdf.gz | 455.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22878_full_validation.pdf.gz emd_22878_full_validation.pdf.gz | 455.2 KB | Display | |
| Data in XML |  emd_22878_validation.xml.gz emd_22878_validation.xml.gz | 7.6 KB | Display | |
| Data in CIF |  emd_22878_validation.cif.gz emd_22878_validation.cif.gz | 9.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22878 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22878 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22878 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10589 (Title: cryo-FIB and cryo-ET study of ribosome and polysome structures in E. coli EMPIAR-10589 (Title: cryo-FIB and cryo-ET study of ribosome and polysome structures in E. coliData size: 9.7 Data #1: Unaligned tilt series of cryo-FIB milling E. coli cells [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22878.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22878.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli polysome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10.91 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ribosome
| Entire | Name: ribosome |
|---|---|
| Components |
|
-Supramolecule #1: ribosome
| Supramolecule | Name: ribosome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)