[English] 日本語
 Yorodumi
Yorodumi- EMDB-22188: CryoEM structure of Chimeric Eastern Equine Encephalitis Virus wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22188 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody | ||||||||||||||||||
 Map data Map data | Eastern Equine Encephalitis Virus (EEEV)_Fab143 complex | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |   Eastern equine encephalitis virus Eastern equine encephalitis virus | ||||||||||||||||||
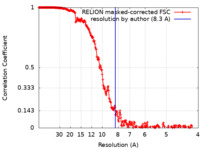
| Method | single particle reconstruction / cryo EM / Resolution: 8.3 Å | ||||||||||||||||||
 Authors Authors | Williamson LE / Gilliland T / Yadav PK / Binshtein E / Bombardi R / Kose N / Nargi RS / Sutton RM / Armstrong E / Carnahan RH ...Williamson LE / Gilliland T / Yadav PK / Binshtein E / Bombardi R / Kose N / Nargi RS / Sutton RM / Armstrong E / Carnahan RH / Walker LM / Kim AS / Fox J / Diamond MS / Ohi M / Klimstra WB / Crowe JE | ||||||||||||||||||
| Funding support |  United States, United Arab Emirates, 5 items United States, United Arab Emirates, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Human Antibodies Protect against Aerosolized Eastern Equine Encephalitis Virus Infection. Authors: Lauren E Williamson / Theron Gilliland / Pramod K Yadav / Elad Binshtein / Robin Bombardi / Nurgun Kose / Rachel S Nargi / Rachel E Sutton / Clarissa L Durie / Erica Armstrong / Robert H ...Authors: Lauren E Williamson / Theron Gilliland / Pramod K Yadav / Elad Binshtein / Robin Bombardi / Nurgun Kose / Rachel S Nargi / Rachel E Sutton / Clarissa L Durie / Erica Armstrong / Robert H Carnahan / Lauren M Walker / Arthur S Kim / Julie M Fox / Michael S Diamond / Melanie D Ohi / William B Klimstra / James E Crowe /  Abstract: Eastern equine encephalitis virus (EEEV) is one of the most virulent viruses endemic to North America. No licensed vaccines or antiviral therapeutics are available to combat this infection, which has ...Eastern equine encephalitis virus (EEEV) is one of the most virulent viruses endemic to North America. No licensed vaccines or antiviral therapeutics are available to combat this infection, which has recently shown an increase in human cases. Here, we characterize human monoclonal antibodies (mAbs) isolated from a survivor of natural EEEV infection with potent (<20 pM) inhibitory activity of EEEV. Cryo-electron microscopy reconstructions of two highly neutralizing mAbs, EEEV-33 and EEEV-143, were solved in complex with chimeric Sindbis/EEEV virions to 7.2 Å and 8.3 Å, respectively. The mAbs recognize two distinct antigenic sites that are critical for inhibiting viral entry into cells. EEEV-33 and EEEV-143 protect against disease following stringent lethal aerosol challenge of mice with highly pathogenic EEEV. These studies provide insight into the molecular basis for the neutralizing human antibody response against EEEV and can facilitate development of vaccines and candidate antibody therapeutics. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22188.map.gz emd_22188.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22188-v30.xml emd-22188-v30.xml emd-22188.xml emd-22188.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22188_fsc.xml emd_22188_fsc.xml | 24.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_22188.png emd_22188.png | 251.8 KB | ||
| Others |  emd_22188_additional_1.map.gz emd_22188_additional_1.map.gz | 146.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22188 http://ftp.pdbj.org/pub/emdb/structures/EMD-22188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22188 | HTTPS FTP |
-Validation report
| Summary document |  emd_22188_validation.pdf.gz emd_22188_validation.pdf.gz | 376.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22188_full_validation.pdf.gz emd_22188_full_validation.pdf.gz | 376.2 KB | Display | |
| Data in XML |  emd_22188_validation.xml.gz emd_22188_validation.xml.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22188 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22188 | HTTPS FTP |
-Related structure data
| Related structure data |  6xo4C  6xobC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10888 (Title: Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-143) EMPIAR-10888 (Title: Motion corrected micrographs - purified SINV/EEEV particles and recombinant anti-EEEV Fab (EEEV-143)Data size: 187.5 Data #1: Motion corrected micrographs Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-143 Antibody [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22188.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22188.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eastern Equine Encephalitis Virus (EEEV)_Fab143 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Eastern Equine Encephalitis Virus (EEEV) Fab143 complex
| File | emd_22188_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Eastern Equine Encephalitis Virus (EEEV)_Fab143 complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern equine encephalitis virus
| Entire | Name:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern equine encephalitis virus
| Supramolecule | Name: Eastern equine encephalitis virus / type: virus / ID: 1 / Parent: 0 / Details: Virus was purified from mammalian cell culture / NCBI-ID: 11021 / Sci species name: Eastern equine encephalitis virus / Sci species strain: Sindbis / Virus type: VIRION / Virus isolate: SUBSPECIES / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Culiseta melanura (mosquito) Culiseta melanura (mosquito) |
| Host system | Organism:  |
| Molecular weight | Theoretical: 6 MDa |
| Virus shell | Shell ID: 1 / Name: Eastern equine encephalitis virus / Diameter: 850.0 Å / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solutions were made fresh | ||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 5 seconds before plunging. | ||||||||||
| Details | Purified SINV/EEEV particles and recombinant Fab mixture |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 77.0 K |
| Details | Preliminary grid screening was performed manually |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 3500 / Average exposure time: 10.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)