[English] 日本語
 Yorodumi
Yorodumi- EMDB-20794: Integrin alpha-v beta-8 in complex with latent TGF-beta (Primary ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20794 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
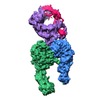
| Title | Integrin alpha-v beta-8 in complex with latent TGF-beta (Primary map: sharpened. Additional map: unsharpened.) | ||||||||||||||||||||||||
 Map data Map data | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation iv (Primary map: sharpened. Additional map: unsharpened.) | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | glycoprotein / adhesion / signaling / SIGNALING PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationPlatelet degranulation / Cell surface interactions at the vascular wall / Molecules associated with elastic fibres / TGF-beta receptor signaling activates SMADs / Syndecan interactions / RUNX3 regulates CDKN1A transcription / RUNX3 regulates p14-ARF / TGFBR3 regulates TGF-beta signaling / Downregulation of TGF-beta receptor signaling / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) ...Platelet degranulation / Cell surface interactions at the vascular wall / Molecules associated with elastic fibres / TGF-beta receptor signaling activates SMADs / Syndecan interactions / RUNX3 regulates CDKN1A transcription / RUNX3 regulates p14-ARF / TGFBR3 regulates TGF-beta signaling / Downregulation of TGF-beta receptor signaling / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / Regulation of RUNX3 expression and activity / ganglioside metabolic process / adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains / positive regulation of microglia differentiation / regulation of interleukin-23 production / branch elongation involved in mammary gland duct branching / positive regulation of primary miRNA processing / columnar/cuboidal epithelial cell maturation / negative regulation of skeletal muscle tissue development / embryonic liver development / regulation of enamel mineralization / regulation of branching involved in mammary gland duct morphogenesis / regulation of cartilage development / regulation of striated muscle tissue development / regulation of protein import into nucleus / tolerance induction to self antigen / extracellular matrix assembly / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of hyaluronan biosynthetic process / type III transforming growth factor beta receptor binding / positive regulation of odontogenesis / Langerhans cell differentiation / negative regulation of macrophage cytokine production / integrin alphav-beta8 complex / integrin alphav-beta6 complex / transforming growth factor beta production / negative regulation of entry of bacterium into host cell / odontoblast differentiation / integrin alphav-beta5 complex / opsonin binding / positive regulation of isotype switching to IgA isotypes / positive regulation of mesenchymal stem cell proliferation / positive regulation of receptor signaling pathway via STAT / membrane protein intracellular domain proteolysis / retina vasculature development in camera-type eye / positive regulation of extracellular matrix assembly / bronchiole development / hyaluronan catabolic process / integrin alphav-beta1 complex / Cross-presentation of particulate exogenous antigens (phagosomes) / mammary gland branching involved in thelarche / extracellular matrix protein binding / positive regulation of vasculature development / lens fiber cell differentiation / ATP biosynthetic process / type II transforming growth factor beta receptor binding / receptor catabolic process / Laminin interactions / placenta blood vessel development / positive regulation of cardiac muscle cell differentiation / integrin alphav-beta3 complex / negative regulation of lipoprotein metabolic process / type I transforming growth factor beta receptor binding / germ cell migration / positive regulation of chemotaxis / entry into host cell by a symbiont-containing vacuole / alphav-beta3 integrin-PKCalpha complex / regulatory T cell differentiation / endoderm development / phospholipid homeostasis / negative regulation of cell-cell adhesion mediated by cadherin / negative regulation of myoblast differentiation / positive regulation of vascular permeability / alphav-beta3 integrin-HMGB1 complex / negative regulation of biomineral tissue development / negative regulation of lipid transport / hard palate development / regulation of phagocytosis / response to cholesterol / oligodendrocyte development / Elastic fibre formation / negative regulation of interleukin-17 production / surfactant homeostasis / cell-cell junction organization / phosphate-containing compound metabolic process / alphav-beta3 integrin-IGF-1-IGF1R complex / transforming growth factor beta binding / deubiquitinase activator activity / positive regulation of small GTPase mediated signal transduction / sprouting angiogenesis / filopodium membrane / extracellular matrix binding / cartilage development / wound healing, spreading of epidermal cells / positive regulation of chemokine (C-X-C motif) ligand 2 production / negative regulation of ossification / apolipoprotein A-I-mediated signaling pathway / negative regulation of low-density lipoprotein particle clearance / aortic valve morphogenesis / apoptotic cell clearance Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||||||||
 Authors Authors | Campbell MG / Cormier A | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Cryo-EM Reveals Integrin-Mediated TGF-β Activation without Release from Latent TGF-β. Authors: Melody G Campbell / Anthony Cormier / Saburo Ito / Robert I Seed / Andrew J Bondesson / Jianlong Lou / James D Marks / Jody L Baron / Yifan Cheng / Stephen L Nishimura /  Abstract: Integrin αvβ8 binds with exquisite specificity to latent transforming growth factor-β (L-TGF-β). This binding is essential for activating L-TGF-β presented by a variety of cell types. ...Integrin αvβ8 binds with exquisite specificity to latent transforming growth factor-β (L-TGF-β). This binding is essential for activating L-TGF-β presented by a variety of cell types. Inhibiting αvβ8-mediated TGF-β activation blocks immunosuppressive regulatory T cell differentiation, which is a potential therapeutic strategy in cancer. Using cryo-electron microscopy, structure-guided mutagenesis, and cell-based assays, we reveal the binding interactions between the entire αvβ8 ectodomain and its intact natural ligand, L-TGF-β, as well as two different inhibitory antibody fragments to understand the structural underpinnings of αvβ8 binding specificity and TGF-β activation. Our studies reveal a mechanism of TGF-β activation where mature TGF-β signals within the confines of L-TGF-β and the release and diffusion of TGF-β are not required. The structural details of this mechanism provide a rational basis for therapeutic strategies to inhibit αvβ8-mediated L-TGF-β activation. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20794.map.gz emd_20794.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20794-v30.xml emd-20794-v30.xml emd-20794.xml emd-20794.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20794.png emd_20794.png | 99.6 KB | ||
| Filedesc metadata |  emd-20794.cif.gz emd-20794.cif.gz | 7.4 KB | ||
| Others |  emd_20794_additional.map.gz emd_20794_additional.map.gz | 52.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20794 http://ftp.pdbj.org/pub/emdb/structures/EMD-20794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20794 | HTTPS FTP |
-Related structure data
| Related structure data |  6ujaMC  6ujbC  6ujcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10343 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a holey carbon grid (strongly preferred orientations) EMPIAR-10343 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a holey carbon grid (strongly preferred orientations)Data size: 1.2 TB Data #1: Unaligned 80-frame movies of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [micrographs - multiframe] Data #2: Dose-weighted aligned micrographs of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [micrographs - single frame] Data #3: Dose-weighted aligned particle stacks of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [picked particles - single frame - processed])  EMPIAR-10344 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a graphene oxide grid (preferred orientations) EMPIAR-10344 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a graphene oxide grid (preferred orientations)Data size: 2.2 TB Data #1: Unaligned 80-frame movies of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [micrographs - multiframe] Data #2: Dose-weighted aligned micrographs of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [micrographs - single frame] Data #3: Dose-weighted aligned particle stacks of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20794.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20794.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation iv (Primary map: sharpened. Additional map: unsharpened.) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.345 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Integrin alpha-v beta-8 in complex with latent TGF-beta,...
| File | emd_20794_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation iv (Additional unsharpened map.) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 ...
| Entire | Name: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 (L-TGF-b1) |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 ...
| Supramolecule | Name: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 (L-TGF-b1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 260 KDa |
-Supramolecule #2: alpha-v beta-8 integrin
| Supramolecule | Name: alpha-v beta-8 integrin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Transforming Growth Factor Beta-1 proprotein
| Supramolecule | Name: Transforming Growth Factor Beta-1 proprotein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 Details: LTGFb is a homodimer in the sample, but only a single chain is modeled. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Integrin alpha-V
| Macromolecule | Name: Integrin alpha-V / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 112.813352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FNLDVDSPAE YSGPEGSYFG FAVDFFVPSA SSRMFLLVGA PKANTTQPGI VEGGQVLKCD WSSTRRCQPI EFDATGNRDY AKDDPLEFK SHQWFGASVR SKQDKILACA PLYHWRTEMK QEREPVGTCF LQDGTKTVEY APCRSQDIDA DGQGFCQGGF S IDFTKADR ...String: FNLDVDSPAE YSGPEGSYFG FAVDFFVPSA SSRMFLLVGA PKANTTQPGI VEGGQVLKCD WSSTRRCQPI EFDATGNRDY AKDDPLEFK SHQWFGASVR SKQDKILACA PLYHWRTEMK QEREPVGTCF LQDGTKTVEY APCRSQDIDA DGQGFCQGGF S IDFTKADR VLLGGPGSFY WQGQLISDQV AEIVSKYDPN VYSIKYNNQL ATRTAQAIFD DSYLGYSVAV GDFNGDGIDD FV SGVPRAA RTLGMVYIYD GKNMSSLYNF TGEQMAAYFG FSVAATDING DDYADVFIGA PLFMDRGSDG KLQEVGQVSV SLQ RASGDF QTTKLNGFEV FARFGSAIAP LGDLDQDGFN DIAIAAPYGG EDKKGIVYIF NGRSTGLNAV PSQILEGQWA ARSM PPSFG YSMKGATDID KNGYPDLIVG AFGVDRAILY RARPVITVNA GLEVYPSILN QDNKTCSLPG TALKVSCFNV RFCLK ADGK GVLPRKLNFQ VELLLDKLKQ KGAIRRALFL YSRSPSHSKN MTISRGGLMQ CEELIAYLRD ESEFRDKLTP ITIFME YRL DYRTAADTTG LQPILNQFTP ANISRQAHIL LDCGEDNVCK PKLEVSVDSD QKKIYIGDDN PLTLIVKAQN QGEGAYE AE LIVSIPLQAD FIGVVRNNEA LARLSCAFKT ENQTRQVVCD LGNPMKAGTQ LLAGLRFSVH QQSEMDTSVK FDLQIQSS N LFDKVSPVVS HKVDLAVLAA VEIRGVSSPD HVFLPIPNWE HKENPETEED VGPVVQHIYE LRNNGPSSFS KAMLHLQWP YKYNNNTLLY ILHYDIDGPM NCTSDMEINP LRIKISSLQT TEKNDTVAGQ GERDHLITKR DLALSEGDIH TLGCGVAQCL KIVCQVGRL DRGKSAILYV KSLLWTETFM NKENQNHSYS LKSSASFNVI EFPYKNLPIE DITNSTLVTT NVTWGIQPAP M PVPVWVII LAVLAGLLLL AVLVFVMYRM GFFKRVRPPQ EEQEREQLQP HENGEGNSET UniProtKB: Integrin alpha-V |
-Macromolecule #2: Integrin beta-8
| Macromolecule | Name: Integrin beta-8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.276664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EDNRCASSNA ASCARCLALG PECGWCVQED FISGGSRSER CDIVSNLISK GCSVDSIEYP SVHVIIPTEN EINTQVTPGE VSIQLRPGA EANFMLKVHP LKKYPVDLYY LVDVSASMHN NIEKLNSVGN DLSRKMAFFS RDFRLGFGSY VDKTVSPYIS I HPERIHNQ ...String: EDNRCASSNA ASCARCLALG PECGWCVQED FISGGSRSER CDIVSNLISK GCSVDSIEYP SVHVIIPTEN EINTQVTPGE VSIQLRPGA EANFMLKVHP LKKYPVDLYY LVDVSASMHN NIEKLNSVGN DLSRKMAFFS RDFRLGFGSY VDKTVSPYIS I HPERIHNQ CSDYNLDCMP PHGYIHVLSL TENITEFEKA VHRQKISGNI DTPEGGFDAM LQAAVCESHI GWRKEAKRLL LV MTDQTSH LALDSKLAGI VVPNDGNCHL KNNVYVKSTT MEHPSLGQLS EKLIDNNINV IFAVQGKQFH WYKDLLPLLP GTI AGEIES KAANLNNLVV EAYQKLISEV KVQVENQVQG IYFNITAICP DGSRKPGMEG CRNVTSNDEV LFNVTVTMKK CDVT GGKNY AIIKPIGFNE TAKIHIHRNC SCQCEDNRGP KGKCVDETFL DSKCFQCDEN KCHFDEDQFS SESCKSHKDQ PVCSG RGVC VCGKCSCHKI KLGKVYGKYC EKDDFSCPYH HGNLCAGHGE CEAGRCQCFS GWEGDRCQCP SAAAQHCVNS KGQVCS GRG TCVCGRCECT DPRSIGRFCE HCPTCYTACK ENWNCMQCLH PHNLSQAILD QCKTSCALME QQHYVDQTSE CFSSPSY LR IFFIIFIVTF LIGLLKVLII RQVILQWNSN KIKSSSDYRV SASKKDKLIL QSVCTRAVTY RREKPEEIKM DISKLNAH E TFRCNF UniProtKB: Integrin beta-8 |
-Macromolecule #3: Transforming growth factor beta-1 proprotein
| Macromolecule | Name: Transforming growth factor beta-1 proprotein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.434449 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPLSTCKTID MELVKRKRIE AIRGQILSKL RLASPPSQGD VPPGPLPEAV LALYNSTRDR VAGESVEPEP EPEADYYAKE VTRVLMVES GNQIYDKFKG TPHSLYMLFN TSELREAVPE PVLLSRAELR LLRLKLKVEQ HVELYQKYSN DSWRYLSNRL L APSDSPEW ...String: GPLSTCKTID MELVKRKRIE AIRGQILSKL RLASPPSQGD VPPGPLPEAV LALYNSTRDR VAGESVEPEP EPEADYYAKE VTRVLMVES GNQIYDKFKG TPHSLYMLFN TSELREAVPE PVLLSRAELR LLRLKLKVEQ HVELYQKYSN DSWRYLSNRL L APSDSPEW LSFDVTGVVR QWLTRREAIE GFRLSAHCSC DSKDNTLHVE INGFNSGRRG DLATIHGMNR PFLLLMATPL ER AQHLHSS RHRRALDTNY CFSSTEKNCC VRQLYIDFRK DLGWKWIHEP KGYHANFCLG PCPYIWSLDT QYSKVLALYN QHN PGASAA PCCVPQALEP LPIVYYVGRK PKVEQLSNMI VRSCKCS UniProtKB: Transforming growth factor beta-1 proprotein |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 5 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 43600 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)