[English] 日本語
 Yorodumi
Yorodumi- EMDB-18842: Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXP... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 1-deoxy-D-xylulose 5-phosphate synthase / thiamin di-phosphate complex / transketolase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information1-deoxy-D-xylulose-5-phosphate synthase / 1-deoxy-D-xylulose-5-phosphate synthase activity / thiamine biosynthetic process / terpenoid biosynthetic process / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
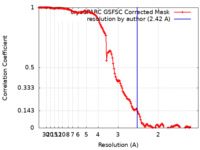
| Method | single particle reconstruction / cryo EM / Resolution: 2.42 Å | |||||||||
 Authors Authors | Gawriljuk VO / Godoy AS / Oerlemans R / Groves MR | |||||||||
| Funding support | European Union,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain. Authors: Victor O Gawriljuk / Andre S Godoy / Rick Oerlemans / Luise A T Welker / Anna K H Hirsch / Matthew R Groves /    Abstract: Plasmodium falciparum is the main causative agent of malaria, a deadly disease that mainly affects children under five years old. Artemisinin-based combination therapies have been pivotal in ...Plasmodium falciparum is the main causative agent of malaria, a deadly disease that mainly affects children under five years old. Artemisinin-based combination therapies have been pivotal in controlling the disease, but resistance has arisen in various regions, increasing the risk of treatment failure. The non-mevalonate pathway is essential for the isoprenoid synthesis in Plasmodium and provides several under-explored targets to be used in the discovery of new antimalarials. 1-deoxy-D-xylulose-5-phosphate synthase (DXPS) is the first and rate-limiting enzyme of the pathway. Despite its importance, there are no structures available for any Plasmodium spp., due to the complex sequence which contains large regions of high disorder, making crystallisation a difficult task. In this manuscript, we use cryo-electron microscopy to solve the P. falciparum DXPS structure at a final resolution of 2.42 Å. Overall, the structure resembles other DXPS enzymes but includes a distinct N-terminal domain exclusive to the Plasmodium genus. Mutational studies show that destabilization of the cap domain interface negatively impacts protein stability and activity. Additionally, a density for the co-factor thiamine diphosphate is found in the active site. Our work highlights the potential of cryo-EM to obtain structures of P. falciparum proteins that are unfeasible by means of crystallography. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18842.map.gz emd_18842.map.gz | 202.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18842-v30.xml emd-18842-v30.xml emd-18842.xml emd-18842.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18842_fsc.xml emd_18842_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18842.png emd_18842.png | 81.2 KB | ||
| Masks |  emd_18842_msk_1.map emd_18842_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18842.cif.gz emd-18842.cif.gz | 6.3 KB | ||
| Others |  emd_18842_additional_1.map.gz emd_18842_additional_1.map.gz emd_18842_half_map_1.map.gz emd_18842_half_map_1.map.gz emd_18842_half_map_2.map.gz emd_18842_half_map_2.map.gz | 121.7 MB 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18842 http://ftp.pdbj.org/pub/emdb/structures/EMD-18842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18842 | HTTPS FTP |
-Related structure data
| Related structure data |  8r2hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18842.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18842.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18842_msk_1.map emd_18842_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
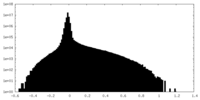
| Density Histograms |
-Additional map: #1
| File | emd_18842_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
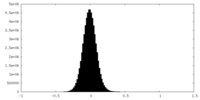
| Density Histograms |
-Half map: #2
| File | emd_18842_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
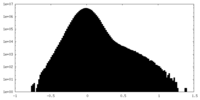
| Density Histograms |
-Half map: #1
| File | emd_18842_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
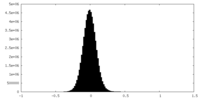
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of 1-deoxy-D-xylulose 5-phosphate synthase with Thiamin dip...
| Entire | Name: Dimer of 1-deoxy-D-xylulose 5-phosphate synthase with Thiamin diphosphate bound. |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of 1-deoxy-D-xylulose 5-phosphate synthase with Thiamin dip...
| Supramolecule | Name: Dimer of 1-deoxy-D-xylulose 5-phosphate synthase with Thiamin diphosphate bound. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 1-deoxy-D-xylulose-5-phosphate synthase
| Macromolecule | Name: 1-deoxy-D-xylulose-5-phosphate synthase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 105.953609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MYDIGKYFKQ INTFINIDEY KTIYGDEIYK EIYELYVERN IPEYYERKYF SEDIKKSVLF DIDKYNDVE FEKAIKEEFI NNGVYINNID NTYYKKENIL IMKKILHYFP LLKLINNPSD LKKLKKQYLP LLAHELKIFL F FIVNITGG ...String: MGSSHHHHHH SSGLVPRGSH MYDIGKYFKQ INTFINIDEY KTIYGDEIYK EIYELYVERN IPEYYERKYF SEDIKKSVLF DIDKYNDVE FEKAIKEEFI NNGVYINNID NTYYKKENIL IMKKILHYFP LLKLINNPSD LKKLKKQYLP LLAHELKIFL F FIVNITGG HFSSVLSSLE IQLLLLYIFN QPYDNVIYDI GHQAYVHKIL TGRKLLFLSL RNKKGISGFL NIFESIYDKF GA GHSSTSL SAIQGYYEAE WQVKNKEKYG NGDIEISDNA NVTNNERIFQ KGIHNDNNIN NNINNNNYIN PSDVVGRENT NVP NVRNDN HNVDKVHIAI IGDGGLTGGM ALEALNYISF LNSKILIIYN DNGQVSLPTN AVSISGNRPI GSISDHLHYF VSNI EANAG DNKLSKNAKE NNIFENLNYD YIGVVNGNNT EELFKVLNNI KENKLKRATV LHVRTKKSND FINSKSPISI LHSIK KNEI FPFDTTILNG NIHKENKIEE EKNVSSSTKY DVNNKNNKNN DNSEIIKYED MFSKETFTDI YTNEMLKYLK KDRNII FLS PAMLGGSGLV KISERYPNNV YDVGIAEQHS VTFAAAMAMN KKLKIQLCIY STFLQRAYDQ IIHDLNLQNI PLKVIIG RS GLVGEDGATH QGIYDLSYLG TLNNAYIISP SNQVDLKRAL RFAYLDKDHS VYIRIPRMNI LSDKYMKGYL NIHMKNES K NIDVNVDIND DVDKYSEEYM DDDNFIKSFI GKSRIIKMDN ENNNTNEHYS SRGDTQTKKK KVCIFNMGSM LFNVINAIK EIEKEQYISH NYSFSIVDMI FLNPLDKNMI DHVIKQNKHQ YLITYEDNTI GGFSTHFNNY LIENNYITKH NLYVHNIYLS NEPIEHASF KDQQEVVKMD KCSLVNRIKN YLKNNPT UniProtKB: 1-deoxy-D-xylulose-5-phosphate synthase |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: THIAMINE DIPHOSPHATE
| Macromolecule | Name: THIAMINE DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: TPP |
|---|---|
| Molecular weight | Theoretical: 425.314 Da |
| Chemical component information |  ChemComp-TPP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 44.47 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)