[English] 日本語
 Yorodumi
Yorodumi- EMDB-18148: a membrane-bound menaquinol:organohalide oxidoreductase complex R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | a membrane-bound menaquinol:organohalide oxidoreductase complex RDH complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RDH menaquinol:organohalide oxidoreductase / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationtetrachloroethene reductive dehalogenase / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) | |||||||||
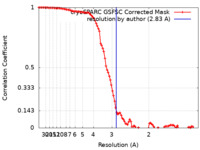
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Dongchun N / Ekundayo B / Henning S / Julien M / Holliger C / Cimmino L | |||||||||
| Funding support |  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of a membrane-bound menaquinol:organohalide oxidoreductase. Authors: Lorenzo Cimmino / Américo G Duarte / Dongchun Ni / Babatunde E Ekundayo / Inês A C Pereira / Henning Stahlberg / Christof Holliger / Julien Maillard /   Abstract: Organohalide-respiring bacteria are key organisms for the bioremediation of soils and aquifers contaminated with halogenated organic compounds. The major players in this process are respiratory ...Organohalide-respiring bacteria are key organisms for the bioremediation of soils and aquifers contaminated with halogenated organic compounds. The major players in this process are respiratory reductive dehalogenases, corrinoid enzymes that use organohalides as substrates and contribute to energy conservation. Here, we present the structure of a menaquinol:organohalide oxidoreductase obtained by cryo-EM. The membrane-bound protein was isolated from Desulfitobacterium hafniense strain TCE1 as a PceAB complex catalysing the dechlorination of tetrachloroethene. Two catalytic PceA subunits are anchored to the membrane by two small integral membrane PceB subunits. The structure reveals two menaquinone molecules bound at the interface of the two different subunits, which are the starting point of a chain of redox cofactors for electron transfer to the active site. In this work, the structure elucidates how energy is conserved during organohalide respiration in menaquinone-dependent organohalide-respiring bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18148.map.gz emd_18148.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18148-v30.xml emd-18148-v30.xml emd-18148.xml emd-18148.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18148_fsc.xml emd_18148_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18148.png emd_18148.png | 63.9 KB | ||
| Filedesc metadata |  emd-18148.cif.gz emd-18148.cif.gz | 5.8 KB | ||
| Others |  emd_18148_half_map_1.map.gz emd_18148_half_map_1.map.gz emd_18148_half_map_2.map.gz emd_18148_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18148 http://ftp.pdbj.org/pub/emdb/structures/EMD-18148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18148 | HTTPS FTP |
-Validation report
| Summary document |  emd_18148_validation.pdf.gz emd_18148_validation.pdf.gz | 189.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18148_full_validation.pdf.gz emd_18148_full_validation.pdf.gz | 189.4 KB | Display | |
| Data in XML |  emd_18148_validation.xml.gz emd_18148_validation.xml.gz | 502 B | Display | |
| Data in CIF |  emd_18148_validation.cif.gz emd_18148_validation.cif.gz | 373 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18148 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18148 | HTTPS FTP |
-Related structure data
| Related structure data |  8q4hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18148.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18148.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18148_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18148_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : a membrane-bound menaquinol:organohalide oxidoreductase
| Entire | Name: a membrane-bound menaquinol:organohalide oxidoreductase |
|---|---|
| Components |
|
-Supramolecule #1: a membrane-bound menaquinol:organohalide oxidoreductase
| Supramolecule | Name: a membrane-bound menaquinol:organohalide oxidoreductase type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 / Details: PceA2B2 |
|---|---|
| Source (natural) | Organism:  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) |
| Molecular weight | Theoretical: 130 kDa/nm |
-Macromolecule #1: Tetrachloroethene reductive dehalogenase
| Macromolecule | Name: Tetrachloroethene reductive dehalogenase / type: protein_or_peptide / ID: 1 / Details: (COB)(SF4)(SF4)(MQ7) / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) |
| Molecular weight | Theoretical: 56.620824 KDa |
| Sequence | String: TSEFPYKVDA KYQRYNSLKN FFEKTFDPEA NKTPIKFHYD DVSKITGKKD TGKDLPTLNA ERLGIKGRPA THTETSILFH TQHLGAMLT QRHNETGWTG LDEALNAGAW AVEFDYSGFN ATGGGPGSVI PLYPINPMTN EIANEPVMVP GLYNWDNIDV E SVRQQGQQ ...String: TSEFPYKVDA KYQRYNSLKN FFEKTFDPEA NKTPIKFHYD DVSKITGKKD TGKDLPTLNA ERLGIKGRPA THTETSILFH TQHLGAMLT QRHNETGWTG LDEALNAGAW AVEFDYSGFN ATGGGPGSVI PLYPINPMTN EIANEPVMVP GLYNWDNIDV E SVRQQGQQ WKFESKEEAS KIVKKATRLL GADLVGIAPY DERWTYSTWG RKIYKPCKMP NGRTKYLPWD LPKMLSGGGV EV FGHAKFE PDWEKYAGFK PKSVIVFVLE EDYEAIRTSP SVISSATVGK SYSNMAEVAY KIAVFLRKLG YYAAPCGNDT GIS VPMAVQ AGLGEAGRNG LLITQKFGPR HRIAKVYTDL ELAPDKPRKF GVREFCRLCK KCADACPAQA ISHEKDPKVL QPED CEVAE NPYTEKWHLD SNRCGSFWAY NGSPCSNCVA VCSWNKVETW NHDVARIATQ IPLLQDAARK FDEWFGYNGP VNPDE RLES GYVQNMVKDF WNNPESIKQ UniProtKB: Tetrachloroethene reductive dehalogenase |
-Macromolecule #2: Probable tetrachloroethene reductive dehalogenase membrane anchor...
| Macromolecule | Name: Probable tetrachloroethene reductive dehalogenase membrane anchor protein type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) |
| Molecular weight | Theoretical: 10.126991 KDa |
| Sequence | String: MNIYDVLIWM ALGMTALLIQ YGIWRYLKGK GKDTIPLQIC GFLANFFFIF ALAWGYSSFS EREYQAIGMG FIFFGGTALI PAIITYRLA UniProtKB: Probable tetrachloroethene reductive dehalogenase membrane anchor protein |
-Macromolecule #3: (~{Z})-1,2-bis(chloranyl)ethene
| Macromolecule | Name: (~{Z})-1,2-bis(chloranyl)ethene / type: ligand / ID: 3 / Number of copies: 2 / Formula: JYF |
|---|---|
| Molecular weight | Theoretical: 96.943 Da |
| Chemical component information | 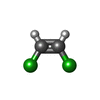 ChemComp-JYF: |
-Macromolecule #4: CO-METHYLCOBALAMIN
| Macromolecule | Name: CO-METHYLCOBALAMIN / type: ligand / ID: 4 / Number of copies: 2 / Formula: COB |
|---|---|
| Molecular weight | Theoretical: 1.344382 KDa |
| Chemical component information |  ChemComp-COB: |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 4 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: MENAQUINONE-7
| Macromolecule | Name: MENAQUINONE-7 / type: ligand / ID: 6 / Number of copies: 2 / Formula: MQ7 |
|---|---|
| Molecular weight | Theoretical: 648.999 Da |
| Chemical component information |  ChemComp-MQ7: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 14 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)