[English] 日本語
 Yorodumi
Yorodumi- PDB-8q4h: a membrane-bound menaquinol:organohalide oxidoreductase complex R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8q4h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | a membrane-bound menaquinol:organohalide oxidoreductase complex RDH complex | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | ELECTRON TRANSPORT / RDH menaquinol:organohalide oxidoreductase | |||||||||
| Function / homology |  Function and homology information Function and homology informationtetrachloroethene reductive dehalogenase / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Dongchun, N. / Ekundayo, B. / Henning, S. / Julien, M. / Holliger, C. / Cimmino, L. | |||||||||
| Funding support |  Switzerland, 2items Switzerland, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of a membrane-bound menaquinol:organohalide oxidoreductase. Authors: Lorenzo Cimmino / Américo G Duarte / Dongchun Ni / Babatunde E Ekundayo / Inês A C Pereira / Henning Stahlberg / Christof Holliger / Julien Maillard /   Abstract: Organohalide-respiring bacteria are key organisms for the bioremediation of soils and aquifers contaminated with halogenated organic compounds. The major players in this process are respiratory ...Organohalide-respiring bacteria are key organisms for the bioremediation of soils and aquifers contaminated with halogenated organic compounds. The major players in this process are respiratory reductive dehalogenases, corrinoid enzymes that use organohalides as substrates and contribute to energy conservation. Here, we present the structure of a menaquinol:organohalide oxidoreductase obtained by cryo-EM. The membrane-bound protein was isolated from Desulfitobacterium hafniense strain TCE1 as a PceAB complex catalysing the dechlorination of tetrachloroethene. Two catalytic PceA subunits are anchored to the membrane by two small integral membrane PceB subunits. The structure reveals two menaquinone molecules bound at the interface of the two different subunits, which are the starting point of a chain of redox cofactors for electron transfer to the active site. In this work, the structure elucidates how energy is conserved during organohalide respiration in menaquinone-dependent organohalide-respiring bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8q4h.cif.gz 8q4h.cif.gz | 224.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8q4h.ent.gz pdb8q4h.ent.gz | 178 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8q4h.json.gz 8q4h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q4/8q4h https://data.pdbj.org/pub/pdb/validation_reports/q4/8q4h ftp://data.pdbj.org/pub/pdb/validation_reports/q4/8q4h ftp://data.pdbj.org/pub/pdb/validation_reports/q4/8q4h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  18148MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 4 molecules ABCD
| #1: Protein | Mass: 56620.824 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: (COB)(SF4)(SF4)(MQ7) Source: (natural)  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria)References: UniProt: Q8GJ31 #2: Protein | Mass: 10126.991 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria)References: UniProt: Q8GJ30 |
|---|
-Non-polymers , 5 types, 24 molecules 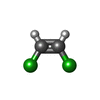








| #3: Chemical | | #4: Chemical | #5: Chemical | ChemComp-SF4 / #6: Chemical | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: a membrane-bound menaquinol:organohalide oxidoreductase Type: COMPLEX / Details: PceA2B2 / Entity ID: #2 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 130 kDa/nm / Experimental value: YES |
| Source (natural) | Organism:  Desulfitobacterium hafniense TCE1 (bacteria) Desulfitobacterium hafniense TCE1 (bacteria) |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
- Processing
Processing
| CTF correction | Type: NONE |
|---|---|
| 3D reconstruction | Resolution: 2.83 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 34078 / Symmetry type: POINT |
 Movie
Movie Controller
Controller


 PDBj
PDBj











