+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
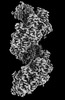
| Title | RAD51 filament on dsDNA bound by the BRCA2 c-terminus | |||||||||
 Map data Map data | RAD51 filament on dsDNA bound by the BRCA2 c-terminus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RAD51 / BRCA2 / Filament / Complex / RECOMBINATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationBRCA2-MAGE-D1 complex / negative regulation of mammary gland epithelial cell proliferation / presynaptic intermediate filament cytoskeleton / mitotic recombination-dependent replication fork processing / chromosome organization involved in meiotic cell cycle / cellular response to camptothecin / establishment of protein localization to telomere / DNA recombinase assembly / telomere maintenance via telomere lengthening / positive regulation of DNA ligation ...BRCA2-MAGE-D1 complex / negative regulation of mammary gland epithelial cell proliferation / presynaptic intermediate filament cytoskeleton / mitotic recombination-dependent replication fork processing / chromosome organization involved in meiotic cell cycle / cellular response to camptothecin / establishment of protein localization to telomere / DNA recombinase assembly / telomere maintenance via telomere lengthening / positive regulation of DNA ligation / Impaired BRCA2 translocation to the nucleus / Impaired BRCA2 binding to SEM1 (DSS1) / double-strand break repair involved in meiotic recombination / nuclear ubiquitin ligase complex / mitotic recombination / DNA strand invasion / cellular response to hydroxyurea / DNA strand exchange activity / lateral element / replication-born double-strand break repair via sister chromatid exchange / telomere maintenance via recombination / histone H4 acetyltransferase activity / histone H3 acetyltransferase activity / regulation of DNA damage checkpoint / Impaired BRCA2 binding to PALB2 / HDR through MMEJ (alt-NHEJ) / gamma-tubulin binding / single-stranded DNA helicase activity / reciprocal meiotic recombination / response to UV-C / DNA repair complex / Homologous DNA Pairing and Strand Exchange / Defective homologous recombination repair (HRR) due to BRCA1 loss of function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA1 binding function / Defective HDR through Homologous Recombination Repair (HRR) due to PALB2 loss of BRCA2/RAD51/RAD51C binding function / Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) / oocyte maturation / Resolution of D-loop Structures through Holliday Junction Intermediates / inner cell mass cell proliferation / DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator / ATP-dependent DNA damage sensor activity / HDR through Single Strand Annealing (SSA) / hematopoietic stem cell proliferation / Impaired BRCA2 binding to RAD51 / regulation of double-strand break repair via homologous recombination / nuclear chromosome / female gonad development / DNA unwinding involved in DNA replication / replication fork processing / male meiosis I / Transcriptional Regulation by E2F6 / Presynaptic phase of homologous DNA pairing and strand exchange / centrosome duplication / response to X-ray / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / ATP-dependent activity, acting on DNA / interstrand cross-link repair / DNA polymerase binding / condensed chromosome / positive regulation of mitotic cell cycle / regulation of cytokinesis / secretory granule / condensed nuclear chromosome / response to gamma radiation / male germ cell nucleus / meiotic cell cycle / cellular response to ionizing radiation / nucleotide-excision repair / double-strand break repair via homologous recombination / brain development / regulation of protein phosphorylation / HDR through Homologous Recombination (HRR) / PML body / Meiotic recombination / cellular senescence / double-strand break repair / site of double-strand break / single-stranded DNA binding / double-stranded DNA binding / spermatogenesis / protease binding / DNA recombination / chromosome, telomeric region / mitochondrial matrix / DNA repair / centrosome / DNA damage response / chromatin binding / regulation of DNA-templated transcription / chromatin / nucleolus / positive regulation of DNA-templated transcription / perinuclear region of cytoplasm / enzyme binding / ATP hydrolysis activity / protein-containing complex / mitochondrion / nucleoplasm / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Appleby R / Pellegrini L | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for stabilisation of the RAD51 nucleoprotein filament by BRCA2. Authors: Robert Appleby / Luay Joudeh / Katie Cobbett / Luca Pellegrini /  Abstract: The BRCA2 tumour suppressor protein preserves genomic integrity via interactions with the DNA-strand exchange RAD51 protein in homology-directed repair. The RAD51-binding TR2 motif at the BRCA2 C- ...The BRCA2 tumour suppressor protein preserves genomic integrity via interactions with the DNA-strand exchange RAD51 protein in homology-directed repair. The RAD51-binding TR2 motif at the BRCA2 C-terminus is essential for protection and restart of stalled replication forks. Biochemical evidence shows that TR2 recognises filamentous RAD51, but existing models of TR2 binding to RAD51 lack a structural basis. Here we used cryo-electron microscopy and structure-guided mutagenesis to elucidate the mechanism of TR2 binding to nucleoprotein filaments of human RAD51. We find that TR2 binds across the protomer interface in the filament, acting as a brace for adjacent RAD51 molecules. TR2 targets an acidic-patch motif on human RAD51 that serves as a recruitment hub in fission yeast Rad51 for recombination mediators Rad52 and Rad55-Rad57. Our findings provide a structural rationale for RAD51 filament stabilisation by BRCA2 and reveal a common recruitment mechanism of recombination mediators to the RAD51 filament. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17585.map.gz emd_17585.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17585-v30.xml emd-17585-v30.xml emd-17585.xml emd-17585.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17585.png emd_17585.png | 76.8 KB | ||
| Filedesc metadata |  emd-17585.cif.gz emd-17585.cif.gz | 6.1 KB | ||
| Others |  emd_17585_half_map_1.map.gz emd_17585_half_map_1.map.gz emd_17585_half_map_2.map.gz emd_17585_half_map_2.map.gz | 23.5 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17585 http://ftp.pdbj.org/pub/emdb/structures/EMD-17585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17585 | HTTPS FTP |
-Validation report
| Summary document |  emd_17585_validation.pdf.gz emd_17585_validation.pdf.gz | 814.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17585_full_validation.pdf.gz emd_17585_full_validation.pdf.gz | 813.9 KB | Display | |
| Data in XML |  emd_17585_validation.xml.gz emd_17585_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_17585_validation.cif.gz emd_17585_validation.cif.gz | 12.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17585 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17585 | HTTPS FTP |
-Related structure data
| Related structure data |  8pbdMC  8pbcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17585.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17585.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RAD51 filament on dsDNA bound by the BRCA2 c-terminus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.304 Å | ||||||||||||||||||||||||||||||||||||
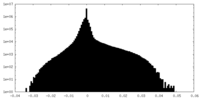
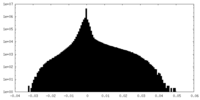
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 1
| File | emd_17585_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
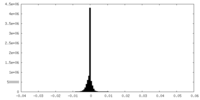
| Density Histograms |
-Half map: Half-map 2
| File | emd_17585_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
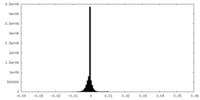
| Density Histograms |
- Sample components
Sample components
+Entire : RAD51 filament formed on dsDNA and bound with a peptide correspon...
+Supramolecule #1: RAD51 filament formed on dsDNA and bound with a peptide correspon...
+Supramolecule #2: DNA repair protein RAD51 homolog 1
+Supramolecule #3: Breast cancer type 2 susceptibility protein
+Supramolecule #4: DNA strand 1 and 2
+Macromolecule #1: DNA repair protein RAD51 homolog 1
+Macromolecule #2: Breast cancer type 2 susceptibility protein
+Macromolecule #3: DNA strand 1
+Macromolecule #4: DNA strand 2
+Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #6: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 15.8 Å Applied symmetry - Helical parameters - Δ&Phi: 56.0 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 2.83 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 1 |
|---|---|
| Startup model | Type of model: OTHER |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION (ver. 3.1) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8pbd: |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)