+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ACADVL dimer from Mus musculus. | ||||||||||||
 Map data Map data | globally sharpened map (locally refined). | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | NADH ubiquinone oxidoreductase / Complex I / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationBeta oxidation of palmitoyl-CoA to myristoyl-CoA / very-long-chain acyl-CoA dehydrogenase / very-long-chain fatty acyl-CoA dehydrogenase activity / negative regulation of fatty acid oxidation / long-chain acyl-CoA dehydrogenase / long-chain fatty acyl-CoA dehydrogenase activity / fatty acid beta-oxidation using acyl-CoA dehydrogenase / fatty-acyl-CoA binding / acyl-CoA dehydrogenase activity / fatty acid catabolic process ...Beta oxidation of palmitoyl-CoA to myristoyl-CoA / very-long-chain acyl-CoA dehydrogenase / very-long-chain fatty acyl-CoA dehydrogenase activity / negative regulation of fatty acid oxidation / long-chain acyl-CoA dehydrogenase / long-chain fatty acyl-CoA dehydrogenase activity / fatty acid beta-oxidation using acyl-CoA dehydrogenase / fatty-acyl-CoA binding / acyl-CoA dehydrogenase activity / fatty acid catabolic process / negative regulation of fatty acid biosynthetic process / regulation of cholesterol metabolic process / fatty acid beta-oxidation / temperature homeostasis / mitochondrial nucleoid / epithelial cell differentiation / response to cold / mitochondrial membrane / flavin adenine dinucleotide binding / mitochondrial inner membrane / nucleolus / mitochondrion / nucleoplasm / identical protein binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
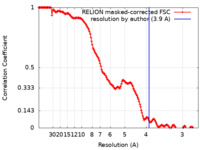
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | ||||||||||||
 Authors Authors | Yin Z / Bridges HR / Agip ANA / Hirst J | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 mouse. Authors: Zhan Yin / Ahmed-Noor A Agip / Hannah R Bridges / Judy Hirst /    Abstract: Respiratory complex I (NADH:ubiquinone oxidoreductase) is essential for cellular energy production and NAD homeostasis. Complex I mutations cause neuromuscular, mitochondrial diseases, such as Leigh ...Respiratory complex I (NADH:ubiquinone oxidoreductase) is essential for cellular energy production and NAD homeostasis. Complex I mutations cause neuromuscular, mitochondrial diseases, such as Leigh Syndrome, but their molecular-level consequences remain poorly understood. Here, we use a popular complex I-linked mitochondrial disease model, the ndufs4 mouse, to define the structural, biochemical, and functional consequences of the absence of subunit NDUFS4. Cryo-EM analyses of the complex I from ndufs4 mouse hearts revealed a loose association of the NADH-dehydrogenase module, and discrete classes containing either assembly factor NDUFAF2 or subunit NDUFS6. Subunit NDUFA12, which replaces its paralogue NDUFAF2 in mature complex I, is absent from all classes, compounding the deletion of NDUFS4 and preventing maturation of an NDUFS4-free enzyme. We propose that NDUFAF2 recruits the NADH-dehydrogenase module during assembly of the complex. Taken together, the findings provide new molecular-level understanding of the ndufs4 mouse model and complex I-linked mitochondrial disease. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16515.map.gz emd_16515.map.gz | 321.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16515-v30.xml emd-16515-v30.xml emd-16515.xml emd-16515.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16515_fsc.xml emd_16515_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16515.png emd_16515.png | 94.7 KB | ||
| Masks |  emd_16515_msk_1.map emd_16515_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16515.cif.gz emd-16515.cif.gz | 5.8 KB | ||
| Others |  emd_16515_half_map_1.map.gz emd_16515_half_map_1.map.gz emd_16515_half_map_2.map.gz emd_16515_half_map_2.map.gz | 321.9 MB 321.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16515 http://ftp.pdbj.org/pub/emdb/structures/EMD-16515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16515 | HTTPS FTP |
-Related structure data
| Related structure data |  8ca1MC  8c2sC  8ca3C  8ca4C  8ca5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16515.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16515.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | globally sharpened map (locally refined). | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.352 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16515_msk_1.map emd_16515_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_16515_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_16515_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
| Entire | Name: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial |
|---|---|
| Components |
|
-Supramolecule #1: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
| Supramolecule | Name: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
| Macromolecule | Name: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: very-long-chain acyl-CoA dehydrogenase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 70.959375 KDa |
| Sequence | String: MQSARMTPSV GRQLLRLGAR SSRSTTVLQG QPRPISAQRL YAREATQAVL DKPETLSSDA STREKPARAE SKSFAVGMFK GQLTIDQVF PYPSVLSEEQ AQFLKELVGP VARFFEEVND PAKNDALEKV EDDTLQGLKE LGAFGLQVPS ELGGLGLSNT Q YARLAEIV ...String: MQSARMTPSV GRQLLRLGAR SSRSTTVLQG QPRPISAQRL YAREATQAVL DKPETLSSDA STREKPARAE SKSFAVGMFK GQLTIDQVF PYPSVLSEEQ AQFLKELVGP VARFFEEVND PAKNDALEKV EDDTLQGLKE LGAFGLQVPS ELGGLGLSNT Q YARLAEIV GMHDLGVSVT LGAHQSIGFK GILLYGTKAQ REKYLPRVAS GQALAAFCLT EPSSGSDVAS IRSSAIPSPC GK YYTLNGS KIWISNGGLA DIFTVFAKTP IKDAATGAVK EKITAFVVER SFGGVTHGLP EKKMGIKASN TSEVYFDGVK VPS ENVLGE VGDGFKVAVN ILNNGRFGMA ATLAGTMKSL IAKAVDHATN RTQFGDKIHN FGVIQEKLAR MAILQYVTES MAYM LSANM DQGFKDFQIE AAISKIFCSE AAWKVADECI QIMGGMGFMK EPGVERVLRD IRIFRIFEGA NDILRLFVAL QGCMD KGKE LTGLGNALKN PFGNVGLLMG EAGKQLRRRT GIGSGLSLSG IVHPELSRSG ELAVQALDQF ATVVEAKLVK HKKGIV NEQ FLLQRLADGA IDLYAMVVVL SRASRSLSEG YPTAQHEKML CDSWCIEAAT RIRENMASLQ SSPQHQELFR NFRSISK AM VENGGLVTGN PLGI UniProtKB: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.82 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.14 Component:
Details: pH was corrected at room temperature ~22 C | |||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)