+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hantaan virus polymerase in replication elongation state | |||||||||
 Map data Map data | Hantaan virus polymerase cryo-EM map in replication elongation state Local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polymerase / replication / bunyavirus / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-templated viral transcription / negative stranded viral RNA replication / cap snatching / endonuclease activity / Hydrolases; Acting on ester bonds / host cell perinuclear region of cytoplasm / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Hantaan virus 76-118 Hantaan virus 76-118 | |||||||||
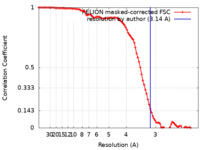
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Durieux trouilleton Q / Arragain B / Malet H | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structures of active Hantaan virus polymerase uncover the mechanisms of Hantaviridae genome replication. Authors: Quentin Durieux Trouilleton / Sergio Barata-García / Benoît Arragain / Juan Reguera / Hélène Malet /  Abstract: Hantaviruses are causing life-threatening zoonotic infections in humans. Their tripartite negative-stranded RNA genome is replicated by the multi-functional viral RNA-dependent RNA-polymerase. Here ...Hantaviruses are causing life-threatening zoonotic infections in humans. Their tripartite negative-stranded RNA genome is replicated by the multi-functional viral RNA-dependent RNA-polymerase. Here we describe the structure of the Hantaan virus polymerase core and establish conditions for in vitro replication activity. The apo structure adopts an inactive conformation that involves substantial folding rearrangement of polymerase motifs. Binding of the 5' viral RNA promoter triggers Hantaan virus polymerase reorganization and activation. It induces the recruitment of the 3' viral RNA towards the polymerase active site for prime-and-realign initiation. The elongation structure reveals the formation of a template/product duplex in the active site cavity concomitant with polymerase core widening and the opening of a 3' viral RNA secondary binding site. Altogether, these elements reveal the molecular specificities of Hantaviridae polymerase structure and uncover the mechanisms underlying replication. They provide a solid framework for future development of antivirals against this group of emerging pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16430.map.gz emd_16430.map.gz | 41.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16430-v30.xml emd-16430-v30.xml emd-16430.xml emd-16430.xml | 26.6 KB 26.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16430_fsc.xml emd_16430_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_16430.png emd_16430.png | 162.8 KB | ||
| Filedesc metadata |  emd-16430.cif.gz emd-16430.cif.gz | 8.2 KB | ||
| Others |  emd_16430_additional_1.map.gz emd_16430_additional_1.map.gz emd_16430_half_map_1.map.gz emd_16430_half_map_1.map.gz emd_16430_half_map_2.map.gz emd_16430_half_map_2.map.gz | 4.9 MB 52 MB 52 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16430 http://ftp.pdbj.org/pub/emdb/structures/EMD-16430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16430 | HTTPS FTP |
-Related structure data
| Related structure data |  8c4vMC  8c4sC  8c4tC  8c4uC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16430.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16430.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hantaan virus polymerase cryo-EM map in replication elongation state Local resolution filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
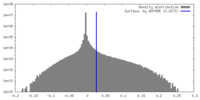
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Hantaan virus polymerase cryo-EM map in replication elongation...
| File | emd_16430_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hantaan virus polymerase cryo-EM map in replication elongation state Post-processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Hantaan virus polymerase cryo-EM map in replication elongation...
| File | emd_16430_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hantaan virus polymerase cryo-EM map in replication elongation state Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Hantaan virus polymerase cryo-EM map in replication elongation...
| File | emd_16430_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hantaan virus polymerase cryo-EM map in replication elongation state Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hantaan virus polymerase in replication elongation state
| Entire | Name: Hantaan virus polymerase in replication elongation state |
|---|---|
| Components |
|
-Supramolecule #1: Hantaan virus polymerase in replication elongation state
| Supramolecule | Name: Hantaan virus polymerase in replication elongation state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Hantaan virus 76-118 / Strain: 76-118 Hantaan virus 76-118 / Strain: 76-118 |
| Molecular weight | Theoretical: 246 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Hantaan virus 76-118 / Strain: 76-118 Hantaan virus 76-118 / Strain: 76-118 |
| Molecular weight | Theoretical: 249.432484 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGHHHHHHDY DIPTTENLYF QGMDKYREIH NKLKEFSPGT LTAVECIDYL DRLYAVRHDI VDQMIKHDWS DNKDSEEAIG KVLLFAGVP SNIITALEKK IIPNHPTGKS LKAFFKMTPA NYKISGTTIE FVEVTVTADV DKGIREKKLK YEAGLTYIEQ E LHKFFLKG ...String: MGHHHHHHDY DIPTTENLYF QGMDKYREIH NKLKEFSPGT LTAVECIDYL DRLYAVRHDI VDQMIKHDWS DNKDSEEAIG KVLLFAGVP SNIITALEKK IIPNHPTGKS LKAFFKMTPA NYKISGTTIE FVEVTVTADV DKGIREKKLK YEAGLTYIEQ E LHKFFLKG EIPQPYKITF NVVAVRTDGS NITTQWPSRR NDGVVQYMRL VQAEISYVRE HLIKTEERAA LEAMFNLKFN IS THKSQPY YIPDYKGMEP IGANIEDLVD YSKDWLSRAR NFSFFEVKGT AVFECFNSNE ANHCQRYPMS RKPRNFLLIQ CSL ITSYKP ATTLSDQIDS RRACSYILNL IPDTPASYLI HDMAYRYINL TREDMINYYA PRIQFKQTQN VREPGTFKLT SSML RAESK AMLDLLNNHK SGEKHGAQIE SLNIASHIVQ SESVSLITKI LSDLELNITE PSTQEYSTTK HTYVDTVLDK FFQNE TQKY LIDVLKKTTA WHIGHLIRDI TESLIAHSGL KRSKYWSLHS YNNGNVILFI LPSKSLEVAG SFIRFITVFR IGPGLV DKD NLDTILIDGD SQWGVSKVMS IDLNRLLALN IAFEKALIAT ATWFQYYTED QGQFPLQYAI RSVFANHFLL AICQKMK LC AIFDNLRYLI PAVTSLYSGF PSLIEKLFER PFKSSLEVYI YYNIKSLLVA LAQNNKARFY SKVKLLGLTV DQSTVGAS G VYPSFMSRIV YKHYRSLISE VTTCFFLFEK GLHGNMNEEA KIHLETVEWA LKFREKEEKY GESLVENGYM MWELRANAE LAEQQLYCQD AIELAAIELN KVLATKSSVV ANSILSKNWE EPYFSQTRNI SLKGMSGQVQ EDGHLSSSVT IIEAIRYLSN SRHNPSLLK LYEETREQKA MARIVRKYQR TEADRGFFIT TLPTRCRLEI IEDYYDAIAK NISEEYISYG GEKKILAIQG A LEKALRWA SGESFIELSN HKFIRMKRKL MYVSADATKW SPGDNSAKFR RFTSMLHNGL PNNKLKNCVI DALKQVYKTD FF MSRKLRN YIDSMESLDP HIKQFLDFFP DGHHGEVKGN WLQGNLNKCS SLFGVAMSLL FKQVWTNLFP ELDCFFEFAH HSD DALFIY GYLEPVDDGT DWFLFVSQQI QAGHLHWFSV NTEMWKSMFN LHEHILLLGS IKISPKKTTV SPTNAEFLST FFEG CAVSI PFVKILLGSL SDLPGLGYFD DLAAAQSRCV KALDLGASPQ VAQLAVALCT SKVERLYGTA PGMVNHPAAY LQVKH TDTP IPLGGNGAMS IMELATAGIG MSDKNLLKRA LLGYSHKRQK SMLYILGLFK FLMKLSDETF QHERLGQFSF IGKVQW KIF TPKSEFEFAD MYTSKFLELW SSQHVTYDYI IPKGRDNLLI YLVRKLNDPS IVTAMTMQSP LQLRFRMQAK QHMKVCR LD GEWVTFREVL AAANSFAENY SATSQDMDLF QTLTSCTFSK EYAWKDFLNG IHCDVIPTKQ VQRAKVARTF TVREKDQI I QNSIPAVIGY KFAVTVEEMS DVLDTAKFPD SLSVDLKTMK DGVYRELGLD ISLPDVMKRI APMLYKSSKS RVVIVQGNV EGTAEAICRY WLKSMSLVKT IRVKPHKEVL QAVSIFNRKE DIGQQKDLAA LKLCIEVWRW CKANSAPYRD WFQALWFEDK TFSEWLDRF CRVGVPPIDP EIQCAALMIA DIKGDYSVLQ LQANRRAYSG KQYDAYCVQT YNEVTKLYEG DLRVTFNFGL D CARLEIFW DKKAYILETS ITQKHVLKIM MDEVSKELIK CGMRFNTEQV QGVRHMVLFK TESGFEWGKP NIPCIVYKNC VL RTSLRTT QAINHKFMIT IKDDGLRAIA QHDEDSPRFL LAHAFHTIRD IRYQAVDAVS NVWFIHKGVK LYLNPIISSG LLE NFMKNL PAAIPPAAYS LIMNRAKISV DLFMFNDLLK LINPRNTLDL SGLETTGDEF STVSSMSSRL WSEEMSLVDD DEEL DDEFT IDLQDVDFEN IDIEADIEHF LQDESSYTGD LLISTEETES KKMRGIVKIL EPVRLIKSWV SRGLSIEKVY SPVNI ILMS RYISKTFNLS TKQVSLLDPY DLTELESIVR GWGECVIDQF ESLDREAQNM VVNKGICPED VIPDSLFSFR HTMVLL RRL FPQDSISSFY UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RNA (5'-R(P*AP*GP*GP*AP*GP*UP*AP*UP*CP*CP*AP*CP*CP*GP*CP*AP*AP*GP...
| Macromolecule | Name: RNA (5'-R(P*AP*GP*GP*AP*GP*UP*AP*UP*CP*CP*AP*CP*CP*GP*CP*AP*AP*GP*A)-3') type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Hantaan virus 76-118 Hantaan virus 76-118 |
| Molecular weight | Theoretical: 6.429919 KDa |
| Sequence | String: UAGGAGUAUC CACCGCAAGA |
-Macromolecule #3: RNA (5'-R(P*AP*CP*UP*AP*CP*UP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*CP*UP*AP*CP*UP*A)-3') / type: rna / ID: 3 / Number of copies: 3 |
|---|---|
| Source (natural) | Organism:  Hantaan virus 76-118 Hantaan virus 76-118 |
| Molecular weight | Theoretical: 7.851604 KDa |
| Sequence | String: CUUUCUUUUG CGGAGUCUAC UACUA |
-Macromolecule #4: RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*A)-3') / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Hantaan virus 76-118 Hantaan virus 76-118 |
| Molecular weight | Theoretical: 8.092932 KDa |
| Sequence | String: UAGUAGUAGA CUCCGCAAAA GAAAG |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: 25 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 276 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3840 pixel / Digitization - Dimensions - Height: 3712 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 3785 / Average exposure time: 5.5 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 2.2 µm / Calibrated magnification: 45454 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 34.7 |
|---|---|
| Output model |  PDB-8c4v: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)