[English] 日本語
 Yorodumi
Yorodumi- EMDB-16373: Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | |||||||||||||||
 Map data Map data | Map generated with SPHIRE Postrefine | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | AAA ATPase / peroxisomes / peroxisome biogenesis / peroxisome biogenesis disorders / Zellweger Syndrome / TRANSLOCASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein import into peroxisome matrix, receptor recycling / protein import into peroxisome matrix / transporter complex / protein transporter activity / peroxisomal membrane / ATPase complex / protein unfolding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / peroxisome / ATP hydrolysis activity ...protein import into peroxisome matrix, receptor recycling / protein import into peroxisome matrix / transporter complex / protein transporter activity / peroxisomal membrane / ATPase complex / protein unfolding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / peroxisome / ATP hydrolysis activity / ATP binding / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||||||||
 Authors Authors | Ruettermann M / Koci M / Lill P / Geladas ED / Kaschani F / Klink BU / Erdmann R / Gatsogiannis C | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate. Authors: Maximilian Rüttermann / Michelle Koci / Pascal Lill / Ermis Dionysios Geladas / Farnusch Kaschani / Björn Udo Klink / Ralf Erdmann / Christos Gatsogiannis /  Abstract: The double-ring AAA+ ATPase Pex1/Pex6 is required for peroxisomal receptor recycling and is essential for peroxisome formation. Pex1/Pex6 mutations cause severe peroxisome associated developmental ...The double-ring AAA+ ATPase Pex1/Pex6 is required for peroxisomal receptor recycling and is essential for peroxisome formation. Pex1/Pex6 mutations cause severe peroxisome associated developmental disorders. Despite its pathophysiological importance, mechanistic details of the heterohexamer are not yet available. Here, we report cryoEM structures of Pex1/Pex6 from Saccharomyces cerevisiae, with an endogenous protein substrate trapped in the central pore of the catalytically active second ring (D2). Pairs of Pex1/Pex6(D2) subdomains engage the substrate via a staircase of pore-1 loops with distinct properties. The first ring (D1) is catalytically inactive but undergoes significant conformational changes resulting in alternate widening and narrowing of its pore. These events are fueled by ATP hydrolysis in the D2 ring and disengagement of a "twin-seam" Pex1/Pex6(D2) heterodimer from the staircase. Mechanical forces are propagated in a unique manner along Pex1/Pex6 interfaces that are not available in homo-oligomeric AAA-ATPases. Our structural analysis reveals the mechanisms of how Pex1 and Pex6 coordinate to achieve substrate translocation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16373.map.gz emd_16373.map.gz | 35.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16373-v30.xml emd-16373-v30.xml emd-16373.xml emd-16373.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16373.png emd_16373.png | 47.9 KB | ||
| Filedesc metadata |  emd-16373.cif.gz emd-16373.cif.gz | 7.9 KB | ||
| Others |  emd_16373_additional_1.map.gz emd_16373_additional_1.map.gz emd_16373_additional_2.map.gz emd_16373_additional_2.map.gz emd_16373_half_map_1.map.gz emd_16373_half_map_1.map.gz emd_16373_half_map_2.map.gz emd_16373_half_map_2.map.gz | 63.9 MB 203.8 MB 36.5 MB 36.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16373 http://ftp.pdbj.org/pub/emdb/structures/EMD-16373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16373 | HTTPS FTP |
-Related structure data
| Related structure data |  8c0wMC  8c0vC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16373.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16373.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map generated with SPHIRE Postrefine | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
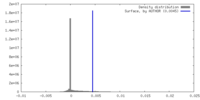
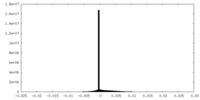
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Density-modified map.
| File | emd_16373_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density-modified map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
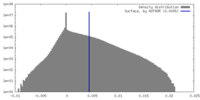
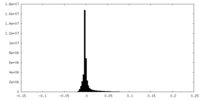
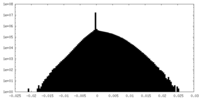
| Density Histograms |
-Additional map: Cryosparc NU-refined map.
| File | emd_16373_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc NU-refined map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
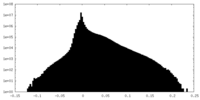
| Density Histograms |
-Half map: Half maps from Relion 3D reconstruction. Input for SPHIRE Postrefine.
| File | emd_16373_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half maps from Relion 3D reconstruction. Input for SPHIRE Postrefine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
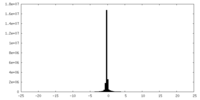
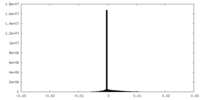
| Density Histograms |
-Half map: Half maps from Relion 3D reconstruction. Input for SPHIRE Postrefine.
| File | emd_16373_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half maps from Relion 3D reconstruction. Input for SPHIRE Postrefine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
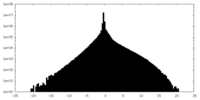
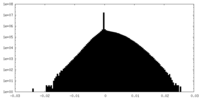
| Density Histograms |
- Sample components
Sample components
-Entire : Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state)
| Entire | Name: Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state) |
|---|---|
| Components |
|
-Supramolecule #1: Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state)
| Supramolecule | Name: Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 774 KDa |
-Supramolecule #2: Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state)
| Supramolecule | Name: Peroxisomal AAA ATPase complex Pex1/Pex6("twin seam" state) type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Unknown peptide
| Supramolecule | Name: Unknown peptide / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Peroxisomal ATPase PEX6
| Macromolecule | Name: Peroxisomal ATPase PEX6 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 115.687094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKASLTFSLS GIYAPCSISR DIYLEYGDKK AECLYGTIRL PQYGPGCTPG KIVHCVLDDS LPFCSIVVPS KLFGFMPTQP TMDFCYFEP ILDNVVPVLD SVTFLINEQL YSKLMDLPQE MQQIQFLHYK YNINSMETVV HSRDILTSGL CQILNCSPFP Q GLVDFTET ...String: MKASLTFSLS GIYAPCSISR DIYLEYGDKK AECLYGTIRL PQYGPGCTPG KIVHCVLDDS LPFCSIVVPS KLFGFMPTQP TMDFCYFEP ILDNVVPVLD SVTFLINEQL YSKLMDLPQE MQQIQFLHYK YNINSMETVV HSRDILTSGL CQILNCSPFP Q GLVDFTET QLILVNDTEQ KLSALKYANE DEEYALPKIG TNSALSIDLE SLPCTISRDL LRPAPHINDD NSIYAFTDAE TL LRLDVTS GSFITVSNMG CVRLVKLFVL LLPNGFKKRT IYAPPKIIAS FPDCSVVTIS KSNIGHTDIP IANQVFISRV GGW LQSQKC FQNIILTTLK KFFSESKRIL CQNDLIPIAF DSSMADLNIA EENDESDDED ELGQYYKNDS LVWFFVTSAE LDCF SKDNS HFIIDPNRTK LITTNITNRR PLPLSRSNLQ RYYGFAETFY YDLHIFPYVR QLVNILETSF NCSQRGITLN ASVLL HSTT NNVGKATMVR FASKYLGIHL LEIDCLSLTS NSRQLDSTSK IIGYIRAKCE NVLPYASPAV IFLAHLDSIL LDVNAN QDP EAIKLQKSIN FEMSKLLDDF TFKFPGTTFV GSVNNIDNVP SSFRSHMRFE ILVPVPSEAQ RLRIFQWYLS SHELNRD VQ QKVPVSYMDN ISFSSLSSYS AGLTPLDIKS IVETARMTAT ARFYQESKKC GWLPQSILIT QEDLSKATSK ARNEFSVS I GAPQIPNVTW DDIGGIDFVK GEILDTIDMP LKHPELFTSG MKKRSGILFY GPPGTGKTLM AKAIATNFSL NFFSVKGPE LLNMYIGESE ANVRRVFQKA REAKPCVIFF DQIDSVAPKR GNQGDSGGVM DRIVSQLLAE LDGMSTDADG VFVIGATNRP DLLDEALLR PGRFDKLLYL GIPDTDTKQL NILEALTRKF VLDNDVKLIE LAKLCPFNYT GADFYALCSD AMLNAMSRIA R MVEKKVSQ HNELTGENIS TRRWFDKIAT KEDTKVVVKM EDFLKAQEQL TPSVSRAELN HYEAVRANFE GA UniProtKB: Peroxisomal ATPase PEX6 |
-Macromolecule #2: Peroxisomal ATPase PEX1
| Macromolecule | Name: Peroxisomal ATPase PEX1 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.758891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TILKNGAIQL LKKVILRSTV CKMDFPKDNL FVVYISDGAQ LPSQKGYASI VKCSLRQSKK SDSDNKSVGI PSKKIGVFIK CDSQIPENH IALSSHLWDA FFTHPMNGAK IKLEFLQMNQ ANIISGRNAT VNIKYFGKDV PTKSGDQYSK LLGGSLLTNN L ILPTEQII ...String: TILKNGAIQL LKKVILRSTV CKMDFPKDNL FVVYISDGAQ LPSQKGYASI VKCSLRQSKK SDSDNKSVGI PSKKIGVFIK CDSQIPENH IALSSHLWDA FFTHPMNGAK IKLEFLQMNQ ANIISGRNAT VNIKYFGKDV PTKSGDQYSK LLGGSLLTNN L ILPTEQII IEIKKGESEQ QLCNLNEISN ESVQWKVTQM GKEEVKDIIE RHLPKHYHVK ETGEVSRTSK DEDDFITVNS IK KEMVNYL TSPIIATPAI ILDGKQGIGK TRLLKELINE VEKDHHIFVK YADCETLHET SNLDKTQKLI MEWCSFCYWY GPS LIVLDN VEALFGKPQA NDGDPSNNGQ WDNASKLLNF FINQVTKIFN KDNKRIRVLF SGKQKTQINP LLFDKHFVSE TWSL RAPDK HARAKLLEYF FSKNQIMKLN RDLQFSDLSL ETEGFSPLDL EIFTEKIFYD LQLERDCDNV VTRELFSKSL SAFTP SALR GVKLTKETNI KWGDIGALAN AKDVLLETLE WPTKYEPIFV NCPLRLRSGI LLYGYPGCGK TLLASAVAQQ CGLNFI SVK GPEILNKFIG ASEQNIRELF ERAQSVKPCI LFFDEFDSIA PKRGHDSTGV TDRVVNQLLT QMDGAEGLDG VYILAAT SR PDLIDSALLR PGRLDKSVIC NIPTESERLD ILQAIVNSKD KDTGQKKFAL EKNADLKLIA EKTAGFSGAD LQGLCYNA Y LKSVHRWLSA ADQSEVVPGN DNIEYFSINE HGRREENRLR LKTLLQQDVV HETKTSTSAA SELTAVVTIN DLLEACQET KPSISTSELV KLRGIYDRFQ KDR UniProtKB: Peroxisomal ATPase PEX1 |
-Macromolecule #3: unknown peptide
| Macromolecule | Name: unknown peptide / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 783.958 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 10 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 10 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK II Details: Grids were blotted for 3.5 sec and plunge-frozen after 1 sec drain time at 100% humidity. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 16763 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8c0w: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)