+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2585 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae Pex1/6 wild type complex bound to ATP | |||||||||
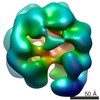
 Map data Map data | negative stain reconstruction of wild type Pex1/6 complex bound to ATP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type-II AAA+ protein complex / heterohexamer / peroxisomal biogenesis | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein import into peroxisome matrix, receptor recycling / protein import into peroxisome matrix / transporter complex / protein transporter activity / peroxisomal membrane / ATPase complex / protein unfolding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / peroxisome / ATP hydrolysis activity ...protein import into peroxisome matrix, receptor recycling / protein import into peroxisome matrix / transporter complex / protein transporter activity / peroxisomal membrane / ATPase complex / protein unfolding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / peroxisome / ATP hydrolysis activity / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Ciniawsky S / Grimm I / Saffian D / Girzalsky W / Erdmann R / Wendler P | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2015 Journal: Nat Commun / Year: 2015Title: Molecular snapshots of the Pex1/6 AAA+ complex in action. Authors: Susanne Ciniawsky / Immanuel Grimm / Delia Saffian / Wolfgang Girzalsky / Ralf Erdmann / Petra Wendler /  Abstract: The peroxisomal proteins Pex1 and Pex6 form a heterohexameric type II AAA+ ATPase complex, which fuels essential protein transport across peroxisomal membranes. Mutations in either ATPase in humans ...The peroxisomal proteins Pex1 and Pex6 form a heterohexameric type II AAA+ ATPase complex, which fuels essential protein transport across peroxisomal membranes. Mutations in either ATPase in humans can lead to severe peroxisomal disorders and early death. We present an extensive structural and biochemical analysis of the yeast Pex1/6 complex. The heterohexamer forms a trimer of Pex1/6 dimers with a triangular geometry that is atypical for AAA+ complexes. While the C-terminal nucleotide-binding domains (D2) of Pex6 constitute the main ATPase activity of the complex, both D2 harbour essential substrate-binding motifs. ATP hydrolysis results in a pumping motion of the complex, suggesting that Pex1/6 function involves substrate translocation through its central channel. Mutation of the Walker B motif in one D2 domain leads to ATP hydrolysis in the neighbouring domain, giving structural insights into inter-domain communication of these unique heterohexameric AAA+ assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2585.map.gz emd_2585.map.gz | 156.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2585-v30.xml emd-2585-v30.xml emd-2585.xml emd-2585.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD_2585.tif EMD_2585.tif | 104.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2585 http://ftp.pdbj.org/pub/emdb/structures/EMD-2585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2585 | HTTPS FTP |
-Validation report
| Summary document |  emd_2585_validation.pdf.gz emd_2585_validation.pdf.gz | 189.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2585_full_validation.pdf.gz emd_2585_full_validation.pdf.gz | 188.7 KB | Display | |
| Data in XML |  emd_2585_validation.xml.gz emd_2585_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2585 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2585 | HTTPS FTP |
-Related structure data
| Related structure data |  2582C  2583C  2584C  2586C  2587C  2588C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2585.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2585.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | negative stain reconstruction of wild type Pex1/6 complex bound to ATP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
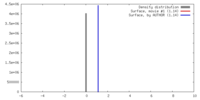
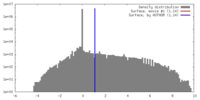
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. cerevisiae Pex1/Pex6 wild type complex
| Entire | Name: S. cerevisiae Pex1/Pex6 wild type complex |
|---|---|
| Components |
|
-Supramolecule #1000: S. cerevisiae Pex1/Pex6 wild type complex
| Supramolecule | Name: S. cerevisiae Pex1/Pex6 wild type complex / type: sample / ID: 1000 Details: heterohexamer purified and assembled in the presence of nucleotide Oligomeric state: heterohexamer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 700 KDa / Theoretical: 700 KDa / Method: gel filtration |
-Macromolecule #1: peroxisomal biogenesis factor 1 (Pex1)
| Macromolecule | Name: peroxisomal biogenesis factor 1 (Pex1) / type: protein_or_peptide / ID: 1 / Name.synonym: Pas1 / Number of copies: 3 / Oligomeric state: heterohexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 117 KDa / Theoretical: 117 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Peroxisomal ATPase PEX1 |
-Macromolecule #2: peroxisomal biogenesis factor 6 (Pex6)
| Macromolecule | Name: peroxisomal biogenesis factor 6 (Pex6) / type: protein_or_peptide / ID: 2 / Name.synonym: Pas8 / Number of copies: 3 / Oligomeric state: heterohexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 116 KDa / Theoretical: 116 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Peroxisomal ATPase PEX6 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris-HCl, 20mM NaCl, 10mM MgCl2, 5mM ATP |
| Staining | Type: NEGATIVE Details: Grid with adsorbed protein was stained with 2% uranyl acetate for 15 seconds and blotted to near dryness. |
| Grid | Details: 400 Cu mesh continuous carbon grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Date | Mar 15, 2013 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Sampling interval: 30 µm / Number real images: 107 / Average electron dose: 18 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.35 µm / Nominal magnification: 103448 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase flipping of micrographs |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: MRC, IMAGIC, SPIDER / Number images used: 1850 |
| Final two d classification | Number classes: 200 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)