[English] 日本語
 Yorodumi
Yorodumi- EMDB-16181: 20S Proteasome subtomogram average from multishot tomography acqu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 20S Proteasome subtomogram average from multishot tomography acquisition on mixed Ribosome-Proteasome sample | |||||||||
 Map data Map data | multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 20s Proteasome / PROTEIN BINDING | |||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Khavnekar S / Plitzko JM / Erdmann PSE | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Multishot tomography for high-resolution in situ subtomogram averaging. Authors: S Khavnekar / W Wan / P Majumder / W Wietrzynski / P S Erdmann / J M Plitzko /    Abstract: Cryo-electron tomography (cryo-ET) and subtomogram averaging (STA) can resolve protein complexes at near atomic resolution, and when combined with focused ion beam (FIB) milling, macromolecules can ...Cryo-electron tomography (cryo-ET) and subtomogram averaging (STA) can resolve protein complexes at near atomic resolution, and when combined with focused ion beam (FIB) milling, macromolecules can be observed within their native context. Unlike single particle acquisition (SPA), cryo-ET can be slow, which may reduce overall project throughput. We here propose a fast, multi-position tomographic acquisition scheme based on beam-tilt corrected beam-shift imaging along the tilt axis, which yields sub-nanometer in situ STA averages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16181.map.gz emd_16181.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16181-v30.xml emd-16181-v30.xml emd-16181.xml emd-16181.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16181.png emd_16181.png | 65 KB | ||
| Masks |  emd_16181_msk_1.map emd_16181_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16181.cif.gz emd-16181.cif.gz | 3.9 KB | ||
| Others |  emd_16181_half_map_1.map.gz emd_16181_half_map_1.map.gz emd_16181_half_map_2.map.gz emd_16181_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16181 http://ftp.pdbj.org/pub/emdb/structures/EMD-16181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16181 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16181 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16181.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
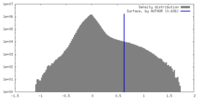
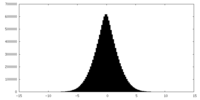
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16181_msk_1.map emd_16181_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
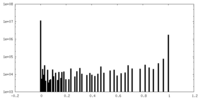
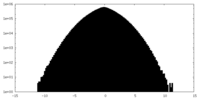
| Density Histograms |
-Half map: multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap
| File | emd_16181_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap | ||||||||||||
| Projections & Slices |
| ||||||||||||
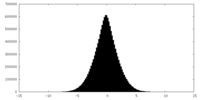
| Density Histograms |
-Half map: multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap
| File | emd_16181_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | multishot tomography Riboprot multishot 2 20S bin1 tomoman-stopgap | ||||||||||||
| Projections & Slices |
| ||||||||||||
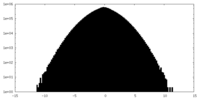
| Density Histograms |
- Sample components
Sample components
-Entire : T. acidophilum 20S Proteasome
| Entire | Name: T. acidophilum 20S Proteasome |
|---|---|
| Components |
|
-Supramolecule #1: T. acidophilum 20S Proteasome
| Supramolecule | Name: T. acidophilum 20S Proteasome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Molecular weight | Theoretical: 750 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: D7 (2x7 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 4.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: STOPGAP / Number subtomograms used: 5000 |
|---|---|
| Extraction | Number tomograms: 25 / Number images used: 5000 / Software - Name: STOPGAP (ver. 0.7) |
| Final 3D classification | Software - Name: STOPGAP |
| Final angle assignment | Type: OTHER / Software - Name: STOPGAP |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)