[English] 日本語
 Yorodumi
Yorodumi- EMDB-14694: Structure of ubiquitinated FANCI in complex with FANCD2 and doubl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fanconi anemia / DNA repair / ubiquitination / ID2 complex / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of CD40 signaling pathway / gamete generation / regulation of regulatory T cell differentiation / double-strand break repair involved in meiotic recombination / homologous chromosome pairing at meiosis / neuronal stem cell population maintenance / brain morphogenesis / response to insecticide / DNA repair complex / mitotic intra-S DNA damage checkpoint signaling ...regulation of CD40 signaling pathway / gamete generation / regulation of regulatory T cell differentiation / double-strand break repair involved in meiotic recombination / homologous chromosome pairing at meiosis / neuronal stem cell population maintenance / brain morphogenesis / response to insecticide / DNA repair complex / mitotic intra-S DNA damage checkpoint signaling / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / interstrand cross-link repair / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / : / condensed chromosome / DNA polymerase binding / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / cytosolic ribosome / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Downregulation of ERBB2:ERBB3 signaling / Regulation of innate immune responses to cytosolic DNA / Pexophagy / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / Regulation of PTEN localization / VLDLR internalisation and degradation / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / positive regulation of protein ubiquitination / Translesion synthesis by REV1 / TICAM1, RIP1-mediated IKK complex recruitment / Regulation of BACH1 activity / Translesion synthesis by POLK / MAP3K8 (TPL2)-dependent MAPK1/3 activation / InlB-mediated entry of Listeria monocytogenes into host cell / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Josephin domain DUBs / Downregulation of TGF-beta receptor signaling / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / response to gamma radiation / IKK complex recruitment mediated by RIP1 / Regulation of activated PAK-2p34 by proteasome mediated degradation / PINK1-PRKN Mediated Mitophagy / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / Autodegradation of Cdh1 by Cdh1:APC/C / TCF dependent signaling in response to WNT / Regulation of NF-kappa B signaling / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / activated TAK1 mediates p38 MAPK activation / TP53 Regulates Transcription of DNA Repair Genes / Asymmetric localization of PCP proteins / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Regulation of signaling by CBL / TNFR2 non-canonical NF-kB pathway / AUF1 (hnRNP D0) binds and destabilizes mRNA Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
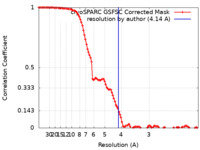
| Method | single particle reconstruction / cryo EM / Resolution: 4.14 Å | |||||||||
 Authors Authors | Lemonidis K / Rennie ML / Arkinson C / Streetley J / Clarke M / Chaugule VK / Walden H | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination. Authors: Kimon Lemonidis / Martin L Rennie / Connor Arkinson / Viduth K Chaugule / Mairi Clarke / James Streetley / Helen Walden /  Abstract: Di-monoubiquitination of the FANCI-FANCD2 (ID2) complex is a central and crucial step for the repair of DNA interstrand crosslinks via the Fanconi anaemia pathway. While FANCD2 ubiquitination ...Di-monoubiquitination of the FANCI-FANCD2 (ID2) complex is a central and crucial step for the repair of DNA interstrand crosslinks via the Fanconi anaemia pathway. While FANCD2 ubiquitination precedes FANCI ubiquitination, FANCD2 is also deubiquitinated at a faster rate than FANCI, which can result in a FANCI-ubiquitinated ID2 complex (I D2). Here, we present a 4.1 Å cryo-EM structure of I D2 complex bound to double-stranded DNA. We show that this complex, like ID2 and I D2 , is also in the closed ID2 conformation and clamps on DNA. The target lysine of FANCD2 (K561) becomes fully exposed in the I D2-DNA structure and is thus primed for ubiquitination. Similarly, FANCI's target lysine (K523) is also primed for ubiquitination in the ID2 -DNA complex. The I D2-DNA complex exhibits deubiquitination resistance, conferred by the presence of DNA and FANCD2. ID2 -DNA, on the other hand, can be efficiently deubiquitinated by USP1-UAF1, unless further ubiquitination on FANCI occurs. Therefore, FANCI ubiquitination effectively maintains FANCD2 ubiquitination in two ways: it prevents excessive FANCD2 deubiquitination within an I D2 -DNA complex, and it enables re-ubiquitination of FANCD2 within a transient, closed-on-DNA, I D2 complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14694.map.gz emd_14694.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14694-v30.xml emd-14694-v30.xml emd-14694.xml emd-14694.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14694_fsc.xml emd_14694_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_14694.png emd_14694.png | 151 KB | ||
| Filedesc metadata |  emd-14694.cif.gz emd-14694.cif.gz | 7.9 KB | ||
| Others |  emd_14694_additional_1.map.gz emd_14694_additional_1.map.gz emd_14694_half_map_1.map.gz emd_14694_half_map_1.map.gz emd_14694_half_map_2.map.gz emd_14694_half_map_2.map.gz | 115.9 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14694 http://ftp.pdbj.org/pub/emdb/structures/EMD-14694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14694 | HTTPS FTP |
-Related structure data
| Related structure data |  7zf1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14694.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14694.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.023 Å | ||||||||||||||||||||||||||||||||||||
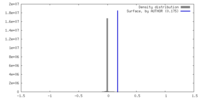
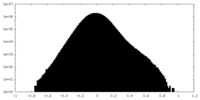
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_14694_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
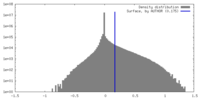
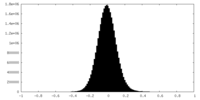
| Density Histograms |
-Half map: #2
| File | emd_14694_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
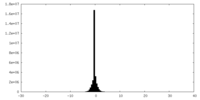
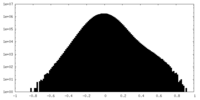
| Density Histograms |
-Half map: #1
| File | emd_14694_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
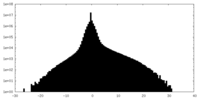
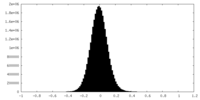
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of ubiquitinated FANCI with FANCD2 and double-stranded DNA.
| Entire | Name: Complex of ubiquitinated FANCI with FANCD2 and double-stranded DNA. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of ubiquitinated FANCI with FANCD2 and double-stranded DNA.
| Supramolecule | Name: Complex of ubiquitinated FANCI with FANCD2 and double-stranded DNA. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fanconi anemia group I protein
| Macromolecule | Name: Fanconi anemia group I protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 152.658594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGKPIPNP LLGLDSTMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKVA GALLRAIFKG SPCSEEAGT LRRRKIYTCC IQLVESGDLQ KEIASEIIGL LMLEAHHFPG PLLVELANEF ISAVREGSLV NGKSLELLPI I LTALATKK ...String: MHHHHHHENL YFQGKPIPNP LLGLDSTMDQ KILSLAAEKT ADKLQEFLQT LREGDLTNLL QNQAVKGKVA GALLRAIFKG SPCSEEAGT LRRRKIYTCC IQLVESGDLQ KEIASEIIGL LMLEAHHFPG PLLVELANEF ISAVREGSLV NGKSLELLPI I LTALATKK ENLAYGKGVL SGEECKKQLI NTLCSGRWDQ QYVIQLTSMF KDVPLTAEEV EFVVEKALSM FSKMNLQEIP PL VYQLLVL SSKGSRKSVL EGIIAFFSAL DKQHNEEQSG DELLDVVTVP SGELRHVEGT IILHIVFAIK LDYELGRELV KHL KVGQQG DSNNNLSPFS IALLLSVTRI QRFQDQVLDL LKTSVVKSFK DLQLLQGSKF LQNLVPHRSY VSTMILEVVK NSVH SWDHV TQGLVELGFI LMDSYGPKKV LDGKTIETSP SLSRMPNQHA CKLGANILLE TFKIHEMIRQ EILEQVLNRV VTRAS SPIS HFLDLLSNIV MYAPLVLQSC SSKVTEAFDY LSFLPLQTVQ RLLKAVQPLL KVSMSMRDCL ILVLRKAMFA NQLDAR KSA VAGFLLLLKN FKVLGSLSSS QCSQSLSVSQ VHVDVHSHYN SVANETFCLE IMDSLRRCLS QQADVRLMLY EGFYDVL RR NSQLANSVMQ TLLSQLKQFY EPKPDLLPPL KLEACILTQG DKISLQEPLD YLLCCIQHCL AWYKNTVIPL QQGEEEEE E EEAFYEDLDD ILESITNRMI KSELEDFELD KSADFSQSTS IGIKNNICAF LVMGVCEVLI EYNFSISSFS KNRFEDILS LFMCYKKLSD ILNEKAGKAK TKMANKTSDS LLSMKFVSSL LTALFRDSIQ SHQESLSVLR SSNEFMRYAV NVALQKVQQL KETGHVSGP DGQNPEKIFQ NLCDITRVLL WRYTSIPTSV EESGKKEKGK SISLLCLEGL QKIFSAVQQF YQPKIQQFLR A LDVTDKEG EEREDADVSV TQRTAFQIRQ FQRSLLNLLS SQEEDFNSKE ALLLVTVLTS LSKLLEPSSP QFVQMLSWTS KI CKENSRE DALFCKSLMN LLFSLHVSYK SPVILLRDLS QDIHGHLGDI DQDVEVEKTN HFAIVNLRTA APTVCLLVLS QAE KVLEEV DWLITKLKGQ VSQETLSEEA SSQATLPNQP VEKAIIMQLG TLLTFFHELV QTALPSGSCV DTLLKDLCKM YTTL TALVR YYLQVCQSSG GIPKNMEKLV KLSGSHLTPL CYSFISYVQN KSKSLNYTGE KKEKPAAVAT AMARVLRETK PIPNL IFAI EQYEKFLIHL SKKSKVNLMQ HMKLSTSRDF KIKGNILDMV LREDGEDENE EGTASEHGGQ NKEPAKKKRK K UniProtKB: Fanconi anemia group I protein |
-Macromolecule #2: Fanconi anemia group D2 protein
| Macromolecule | Name: Fanconi anemia group D2 protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 166.313719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSMVS KRRLSKSEDK ESLTEDASKT RKQPLSKKTK KSHIANEVEE NDSIFVKLLK ISGIILKTGE SQNQLAVDQ IAFQKKLFQT LRRHPSYPKI IEEFVSGLES YIEDEDSFRN CLLSCERLQD EEASMGASYS KSLIKLLLGI D ILQPAIIK ...String: MHHHHHHLEV LFQGPGSMVS KRRLSKSEDK ESLTEDASKT RKQPLSKKTK KSHIANEVEE NDSIFVKLLK ISGIILKTGE SQNQLAVDQ IAFQKKLFQT LRRHPSYPKI IEEFVSGLES YIEDEDSFRN CLLSCERLQD EEASMGASYS KSLIKLLLGI D ILQPAIIK TLFEKLPEYF FENKNSDEIN IPRLIVSQLK WLDRVVDGKD LTTKIMQLIS IAPENLQHDI ITSLPEILGD SQ HADVGKE LSDLLIENTS LTVPILDVLS SLRLDPNFLL KVRQLVMDKL SSIRLEDLPV IIKFILHSVT AMDTLEVISE LRE KLDLQH CVLPSRLQAS QVKLKSKGRA SSSGNQESSG QSCIILLFDV IKSAIRYEKT ISEAWIKAIE NTASVSEHKV FDLV MLFII YSTNTQTKKY IDRVLRNKIR SGCIQEQLLQ STFSVHYLVL KDMCSSILSL AQSLLHSLDQ SIISFGSLLY KYAFK FFDT YCQQEVVGAL VTHICSGNEA EVDTALDVLL ELVVLNPSAM MMNAVFVKGI LDYLDNISPQ QIRKLFYVLS TLAFSK QNE ASSHIQDDMH LVIRKQLSST VFKYKLIGII GAVTMAGIMA ADRSESPSLT QERANLSDEQ CTQVTSLLQL VHSCSEQ SP QASALYYDEF ANLIQHEKLD PKALEWVGHT ICNDFQDAFV VDSCVVPEGD FPFPVKALYG LEEYDTQDGI AINLLPLL F SQDFAKDGGP VTSQESGQKL VSPLCLAPYF RLLRLCVERQ HNGNLEEIDG LLDCPIFLTD LEPGEKLESM SAKERSFMC SLIFLTLNWF REIVNAFCQE TSPEMKGKVL TRLKHIVELQ IILEKYLAVT PDYVPPLGNF DVETLDITPH TVTAISAKIR KKGKIERKQ KTDGSKTSSS DTLSEEKNSE CDPTPSHRGQ LNKEFTGKEE KTSLLLHNSH AFFRELDIEV FSILHCGLVT K FILDTEMH TEATEVVQLG PPELLFLLED LSQKLESMLT PPIARRVPFL KNKGSRNIGF SHLQQRSAQE IVHCVFQLLT PM CNHLENI HNYFQCLAAE NHGVVDGPGV KVQEYHIMSS CYQRLLQIFH GLFAWSGFSQ PENQNLLYSA LHVLSSRLKQ GEH SQPLEE LLSQSVHYLQ NFHQSIPSFQ CALYLIRLLM VILEKSTASA QNKEKIASLA RQFLCRVWPS GDKEKSNISN DQLH ALLCI YLEHTESILK AIEEIAGVGV PELINSPKDA SSSTFPTLTR HTFVVFFRVM MAELEKTVKK IEPGTAADSQ QIHEE KLLY WNMAVRDFSI LINLIKVFDS HPVLHVCLKY GRLFVEAFLK QCMPLLDFSF RKHREDVLSL LETFQLDTRL LHHLCG HSK IHQDTRLTQH VPLLKKTLEL LVCRVKAMLT LNNCREAFWL GNLKNRDLQG EEIKSQNSQE STADESEDDM SSQASKS KA TEDGEEDEVS AGEKEQDSDE SYDDSD UniProtKB: Fanconi anemia group D2 protein |
-Macromolecule #3: Ubiquitin-60S ribosomal protein L40
| Macromolecule | Name: Ubiquitin-60S ribosomal protein L40 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.875125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMQIFVK TLTGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL SDYNIQKEST LHLVLRLRGG UniProtKB: Ubiquitin-ribosomal protein eL40 fusion protein |
-Macromolecule #4: DNA (61-MER)
| Macromolecule | Name: DNA (61-MER) / type: dna / ID: 4 Details: 61-MER DNA modelled using chain S of PDB entry 6VAE as initial model for refinement. Complementary to chain S. Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.951746 KDa |
| Sequence | String: (DG)(DG)(DC)(DA)(DC)(DA)(DG)(DG)(DT)(DT) (DC)(DA)(DG)(DA)(DG)(DC)(DA)(DG)(DG)(DC) (DG)(DT)(DT)(DC)(DC)(DG)(DT)(DT)(DC) |
-Macromolecule #5: DNA (61-MER)
| Macromolecule | Name: DNA (61-MER) / type: dna / ID: 5 Details: 61-MER DNA modelled using chain T of PDB entry 6VAE as initial model for refinement. Complementary to chain S. Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.880711 KDa |
| Sequence | String: (DG)(DA)(DA)(DC)(DG)(DG)(DA)(DA)(DC)(DG) (DC)(DC)(DT)(DG)(DC)(DT)(DC)(DT)(DG)(DA) (DA)(DC)(DC)(DT)(DG)(DT)(DG)(DC)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Detector mode: COUNTING / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 0.5 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-7zf1: |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)