[English] 日本語
 Yorodumi
Yorodumi- EMDB-14527: Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 che... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | |||||||||
 Map data Map data | Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA damage checkpoint / Rad17-RFC / 9-1-1 / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmeiotic DNA integrity checkpoint signaling / checkpoint clamp complex / meiotic recombination checkpoint signaling / Rad17 RFC-like complex / Elg1 RFC-like complex / DNA replication factor C complex / Ctf18 RFC-like complex / regulation of phosphorylation / exodeoxyribonuclease III / DNA clamp loader activity ...meiotic DNA integrity checkpoint signaling / checkpoint clamp complex / meiotic recombination checkpoint signaling / Rad17 RFC-like complex / Elg1 RFC-like complex / DNA replication factor C complex / Ctf18 RFC-like complex / regulation of phosphorylation / exodeoxyribonuclease III / DNA clamp loader activity / double-stranded DNA 3'-5' DNA exonuclease activity / Polymerase switching / positive regulation of DNA-directed DNA polymerase activity / DNA replication checkpoint signaling / embryo development ending in birth or egg hatching / mitotic DNA replication checkpoint signaling / protein localization to site of double-strand break / Polymerase switching on the C-strand of the telomere / chromatin-protein adaptor activity / mitotic intra-S DNA damage checkpoint signaling / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage / HDR through Single Strand Annealing (SSA) / DNA strand elongation involved in DNA replication / DNA synthesis involved in DNA repair / Impaired BRCA2 binding to RAD51 / negative regulation of DNA replication / Presynaptic phase of homologous DNA pairing and strand exchange / PCNA-Dependent Long Patch Base Excision Repair / ATP-dependent activity, acting on DNA / Activation of ATR in response to replication stress / response to UV / 3'-5' exonuclease activity / substantia nigra development / telomere maintenance / DNA damage checkpoint signaling / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / cellular response to ionizing radiation / nucleotide-excision repair / Termination of translesion DNA synthesis / Translesion Synthesis by POLH / Recognition of DNA damage by PCNA-containing replication complex / double-strand break repair via homologous recombination / G2/M DNA damage checkpoint / SH3 domain binding / HDR through Homologous Recombination (HRR) / DNA-templated DNA replication / Dual Incision in GG-NER / histone deacetylase binding / intrinsic apoptotic signaling pathway in response to DNA damage / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / site of double-strand break / chromosome / Processing of DNA double-strand break ends / Regulation of TP53 Activity through Phosphorylation / damaged DNA binding / DNA replication / intracellular membrane-bounded organelle / DNA repair / DNA damage response / chromatin binding / protein kinase binding / nucleolus / enzyme binding / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
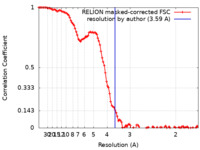
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Day M / Oliver AW / Pearl LH | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction. Authors: Matthew Day / Antony W Oliver / Laurence H Pearl /  Abstract: The RAD9-RAD1-HUS1 (9-1-1) clamp forms one half of the DNA damage checkpoint system that signals the presence of substantial regions of single-stranded DNA arising from replication fork collapse or ...The RAD9-RAD1-HUS1 (9-1-1) clamp forms one half of the DNA damage checkpoint system that signals the presence of substantial regions of single-stranded DNA arising from replication fork collapse or resection of DNA double strand breaks. Loaded at the 5'-recessed end of a dsDNA-ssDNA junction by the RAD17-RFC clamp loader complex, the phosphorylated C-terminal tail of the RAD9 subunit of 9-1-1 engages with the mediator scaffold TOPBP1 which in turn activates the ATR kinase, localised through the interaction of its constitutive partner ATRIP with RPA-coated ssDNA. Using cryogenic electron microscopy (cryoEM) we have determined the structure of a complex of the human RAD17-RFC clamp loader bound to human 9-1-1, engaged with a dsDNA-ssDNA junction. The structure answers the key questions of how RAD17 confers specificity for 9-1-1 over PCNA, and how the clamp loader specifically recognises the recessed 5' DNA end and fixes the orientation of 9-1-1 on the ssDNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14527.map.gz emd_14527.map.gz | 87.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14527-v30.xml emd-14527-v30.xml emd-14527.xml emd-14527.xml | 28.6 KB 28.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14527_fsc.xml emd_14527_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14527.png emd_14527.png | 113.7 KB | ||
| Filedesc metadata |  emd-14527.cif.gz emd-14527.cif.gz | 9 KB | ||
| Others |  emd_14527_half_map_1.map.gz emd_14527_half_map_1.map.gz emd_14527_half_map_2.map.gz emd_14527_half_map_2.map.gz | 77.4 MB 77.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14527 http://ftp.pdbj.org/pub/emdb/structures/EMD-14527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14527 | HTTPS FTP |
-Related structure data
| Related structure data |  7z6hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11062 (Title: Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction EMPIAR-11062 (Title: Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junctionData size: 4.4 TB Data #1: unaligned multiframe EER movies for human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14527.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14527.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
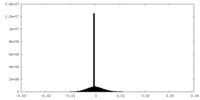
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 1
| File | emd_14527_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
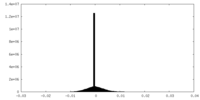
| Projections & Slices |
| ||||||||||||
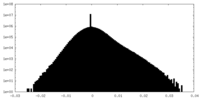
| Density Histograms |
-Half map: Half-map 2
| File | emd_14527_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
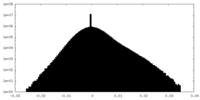
| Density Histograms |
- Sample components
Sample components
+Entire : DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp
+Supramolecule #1: DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp
+Macromolecule #1: Cell cycle checkpoint control protein RAD9A
+Macromolecule #2: Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17
+Macromolecule #3: Checkpoint protein HUS1
+Macromolecule #4: Replication factor C subunit 4
+Macromolecule #5: Replication factor C subunit 3
+Macromolecule #6: Replication factor C subunit 2
+Macromolecule #7: Replication factor C subunit 5
+Macromolecule #8: Hairpin DNA
+Macromolecule #9: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.21 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 34.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)