[English] 日本語
 Yorodumi
Yorodumi- EMDB-13322: Peripheral row of the protein scaffold at rod outer segment disk ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13322 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Peripheral row of the protein scaffold at rod outer segment disk rims in wild type mice (conventional defocused data). | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
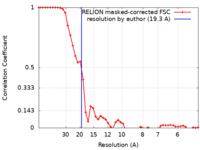
| Method | subtomogram averaging / cryo EM / Resolution: 19.3 Å | |||||||||
 Authors Authors | Poege M / Mahamid J / Imanishi SS / Plitzko JM / Palczewski K / Baumeister W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Determinants shaping the nanoscale architecture of the mouse rod outer segment. Authors: Matthias Pöge / Julia Mahamid / Sanae S Imanishi / Jürgen M Plitzko / Krzysztof Palczewski / Wolfgang Baumeister /   Abstract: The unique membrane organization of the rod outer segment (ROS), the specialized sensory cilium of rod photoreceptor cells, provides the foundation for phototransduction, the initial step in vision. ...The unique membrane organization of the rod outer segment (ROS), the specialized sensory cilium of rod photoreceptor cells, provides the foundation for phototransduction, the initial step in vision. ROS architecture is characterized by a stack of identically shaped and tightly packed membrane disks loaded with the visual receptor rhodopsin. A wide range of genetic aberrations have been reported to compromise ROS ultrastructure, impairing photoreceptor viability and function. Yet, the structural basis giving rise to the remarkably precise arrangement of ROS membrane stacks and the molecular mechanisms underlying genetically inherited diseases remain elusive. Here, cryo-electron tomography (cryo-ET) performed on native ROS at molecular resolution provides insights into key structural determinants of ROS membrane architecture. Our data confirm the existence of two previously observed molecular connectors/spacers which likely contribute to the nanometer-scale precise stacking of the ROS disks. We further provide evidence that the extreme radius of curvature at the disk rims is enforced by a continuous supramolecular assembly composed of peripherin-2 (PRPH2) and rod outer segment membrane protein 1 (ROM1) oligomers. We suggest that together these molecular assemblies constitute the structural basis of the highly specialized ROS functional architecture. Our Cryo-ET data provide novel quantitative and structural information on the molecular architecture in ROS and substantiate previous results on proposed mechanisms underlying pathologies of certain PRPH2 mutations leading to blindness. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13322.map.gz emd_13322.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13322-v30.xml emd-13322-v30.xml emd-13322.xml emd-13322.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13322_fsc.xml emd_13322_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13322.png emd_13322.png | 47.6 KB | ||
| Masks |  emd_13322_msk_1.map emd_13322_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_13322_additional_1.map.gz emd_13322_additional_1.map.gz emd_13322_half_map_1.map.gz emd_13322_half_map_1.map.gz emd_13322_half_map_2.map.gz emd_13322_half_map_2.map.gz | 1.5 MB 5.9 MB 5.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13322 http://ftp.pdbj.org/pub/emdb/structures/EMD-13322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13322 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13322 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10773 (Title: Cryo-electron tomography of rod outer segments in wild type mice acquired conventionally with defocus EMPIAR-10773 (Title: Cryo-electron tomography of rod outer segments in wild type mice acquired conventionally with defocusData size: 4.1 Data #1: Tomograms of rod outer segments in wild type mice acquired conventionally with defocus [reconstructed volumes]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13322.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13322.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13322_msk_1.map emd_13322_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Masked volume focusing on 4 repeats along the...
| File | emd_13322_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked volume focusing on 4 repeats along the peripheral row with its connections to the central row. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map one as output of RELION Refine3D.
| File | emd_13322_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map one as output of RELION Refine3D. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map two as output of RELION Refine3D.
| File | emd_13322_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map two as output of RELION Refine3D. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rod outer segment disk rim scaffold
| Entire | Name: Rod outer segment disk rim scaffold |
|---|---|
| Components |
|
-Supramolecule #1: Rod outer segment disk rim scaffold
| Supramolecule | Name: Rod outer segment disk rim scaffold / type: complex / ID: 1 / Parent: 0 Details: Protein scaffold located at the outer periphery of rod outer segment disk rims in wild type mice - the peripheral row. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: Ringers buffer |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 310 K / Instrument: FEI VITROBOT MARK IV |
| Details | Rod outer segments were isolated by retinal detachment with a single physical disruption, applied to EM-grids and plunge frozen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.8 e/Å2 Details: The total accumulated dose per tilt series was ~ 100e-/A^2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)