[English] 日本語
 Yorodumi
Yorodumi- EMDB-11708: Cryo-EM map of the activated B. subtilis ClpC arranged as a tetra... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11708 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
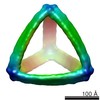
| Title | Cryo-EM map of the activated B. subtilis ClpC arranged as a tetramer of hexamers | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationestablishment of competence for transformation / ATP hydrolysis activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Morreale FE / Meinhart A / Haselbach D / Clausen T | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: BacPROTACs mediate targeted protein degradation in bacteria. Authors: Francesca E Morreale / Stefan Kleine / Julia Leodolter / Sabryna Junker / David M Hoi / Stepan Ovchinnikov / Anastasia Okun / Juliane Kley / Robert Kurzbauer / Lukas Junk / Somraj Guha / ...Authors: Francesca E Morreale / Stefan Kleine / Julia Leodolter / Sabryna Junker / David M Hoi / Stepan Ovchinnikov / Anastasia Okun / Juliane Kley / Robert Kurzbauer / Lukas Junk / Somraj Guha / David Podlesainski / Uli Kazmaier / Guido Boehmelt / Harald Weinstabl / Klaus Rumpel / Volker M Schmiedel / Markus Hartl / David Haselbach / Anton Meinhart / Markus Kaiser / Tim Clausen /   Abstract: Hijacking the cellular protein degradation system offers unique opportunities for drug discovery, as exemplified by proteolysis-targeting chimeras. Despite their great promise for medical chemistry, ...Hijacking the cellular protein degradation system offers unique opportunities for drug discovery, as exemplified by proteolysis-targeting chimeras. Despite their great promise for medical chemistry, so far, it has not been possible to reprogram the bacterial degradation machinery to interfere with microbial infections. Here, we develop small-molecule degraders, so-called BacPROTACs, that bind to the substrate receptor of the ClpC:ClpP protease, priming neo-substrates for degradation. In addition to their targeting function, BacPROTACs activate ClpC, transforming the resting unfoldase into its functional state. The induced higher-order oligomer was visualized by cryo-EM analysis, providing a structural snapshot of activated ClpC unfolding a protein substrate. Finally, drug susceptibility and degradation assays performed in mycobacteria demonstrate in vivo activity of BacPROTACs, allowing selective targeting of endogenous proteins via fusion to an established degron. In addition to guiding antibiotic discovery, the BacPROTAC technology presents a versatile research tool enabling the inducible degradation of bacterial proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11708.map.gz emd_11708.map.gz | 445.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11708-v30.xml emd-11708-v30.xml emd-11708.xml emd-11708.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11708.png emd_11708.png | 88.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11708 http://ftp.pdbj.org/pub/emdb/structures/EMD-11708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11708 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11708 | HTTPS FTP |
-Related structure data
| Related structure data |  7aa4C  7abrC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11068 (Title: Single particle Data of the activated B. subtilis ClpC arranged as a tetramer of hexamers EMPIAR-11068 (Title: Single particle Data of the activated B. subtilis ClpC arranged as a tetramer of hexamersData size: 4.9 TB Data #1: Unaligned Multiframe micrographs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11708.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11708.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative regulator of genetic competence ClpC/MecB
| Entire | Name: Negative regulator of genetic competence ClpC/MecB |
|---|---|
| Components |
|
-Supramolecule #1: Negative regulator of genetic competence ClpC/MecB
| Supramolecule | Name: Negative regulator of genetic competence ClpC/MecB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Negative regulator of genetic competence ClpC/MecB
| Macromolecule | Name: Negative regulator of genetic competence ClpC/MecB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MMFGRFTERA QKVLALAQEE ALRLGHNNIG TEHILLGLVR EGEGIAAKAL QALGLGSEKI QKEVESLIGR GQEMSQTIHY TPRAKKVIE LSMDEARKLG HSYVGTEHIL LGLIREGEGV AARVLNNLGV SLNKARQQVL QLLGSNETGS SAAGTNSNAN T PTLDSLAR ...String: MMFGRFTERA QKVLALAQEE ALRLGHNNIG TEHILLGLVR EGEGIAAKAL QALGLGSEKI QKEVESLIGR GQEMSQTIHY TPRAKKVIE LSMDEARKLG HSYVGTEHIL LGLIREGEGV AARVLNNLGV SLNKARQQVL QLLGSNETGS SAAGTNSNAN T PTLDSLAR DLTAIAKEDS LDPVIGRSKE IQRVIEVLSR RTKNNPVLIG EPGVGKTAIA EGLAQQIINN EVPEILRDKR VM TLDMGTV VAGTKYRGEF EDRLKKVMDE IRQAGNIILF IDALHTLIGA GGAEGAIDAS NILKPSLARG ELQCIGATTL DEY RKYIEK DAALERRFQP IQVDQPSVDE SIQILQGLRD RYEAHHRVSI TDDAIEAAVK LSDRYISDRF LPDKAIDLID EAGS KVRLR SFTTPPNLKE LEQKLDEVRK EKDAAVQSQE FEKAASLRDT EQRLREQVED TKKSWKEKQG QENSEVTVDD IAMVV SSWT GVPVSKIAQT ETDKLLNMEN ILHSRVIGQD EAVVAVAKAV RRARAGLKDP KRPIGSFIFL GPTGVGKTEL ARALAE SIF GDEESMIRID MSEYMEKHST SRLVGSPPGY VGYDEGGQLT EKVRRKPYSV VLLDAIEKAH PDVFNILLQV LEDGRLT DS KGRTVDFRNT ILIMTSNVGA SELKRNKYVG FNVQDETQNH KDMKDKVMGE LKRAFRPEFI NRIDEIIVFH SLEKKHLT E IVSLMSDQLT KRLKEQDLSI ELTDAAKAKV AEEGVDLEYG ARPLRRAIQK HVEDRLSEEL LRGNIHKGQH IVLDVEDGE FVVKTTAKTN LEHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 4455 / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















