[English] 日本語
 Yorodumi
Yorodumi- EMDB-0122: Cryo-EM structure of the hub of the 32 triskelia sweet potato cla... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0122 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the hub of the 32 triskelia sweet potato clathrin coat complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.79 Å | |||||||||
 Authors Authors | Morris KL / Smith CJ | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly. Authors: Kyle L Morris / Joseph R Jones / Mary Halebian / Shenping Wu / Michael Baker / Jean-Paul Armache / Amaurys Avila Ibarra / Richard B Sessions / Alexander D Cameron / Yifan Cheng / Corinne J Smith /   Abstract: Clathrin forms diverse lattice and cage structures that change size and shape rapidly in response to the needs of eukaryotic cells during clathrin-mediated endocytosis and intracellular trafficking. ...Clathrin forms diverse lattice and cage structures that change size and shape rapidly in response to the needs of eukaryotic cells during clathrin-mediated endocytosis and intracellular trafficking. We present the cryo-EM structure and molecular model of assembled porcine clathrin, providing insights into interactions that stabilize key elements of the clathrin lattice, namely, between adjacent heavy chains, at the light chain-heavy chain interface and within the trimerization domain. Furthermore, we report cryo-EM maps for five different clathrin cage architectures. Fitting structural models to three of these maps shows that their assembly requires only a limited range of triskelion leg conformations, yet inherent flexibility is required to maintain contacts. Analysis of the protein-protein interfaces shows remarkable conservation of contact sites despite architectural variation. These data reveal a universal mode of clathrin assembly that allows variable cage architecture and adaptation of coated vesicle size and shape during clathrin-mediated vesicular trafficking or endocytosis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0122.map.gz emd_0122.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0122-v30.xml emd-0122-v30.xml emd-0122.xml emd-0122.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0122_fsc.xml emd_0122_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0122.png emd_0122.png | 113.6 KB | ||
| Masks |  emd_0122_msk_1.map emd_0122_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_0122_additional.map.gz emd_0122_additional.map.gz emd_0122_additional_1.map.gz emd_0122_additional_1.map.gz emd_0122_half_map_1.map.gz emd_0122_half_map_1.map.gz emd_0122_half_map_2.map.gz emd_0122_half_map_2.map.gz | 8.1 MB 8.1 MB 15 MB 15 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0122 http://ftp.pdbj.org/pub/emdb/structures/EMD-0122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0122 | HTTPS FTP |
-Related structure data
| Related structure data |  0114C  0115C  0116C  0118C  0120C  0121C  0123C  0124C  0125C  0126C  6sctC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10296 (Title: Single particle cryo-EM dataset of the triskelion hub subparticle extraction from clathrin cages EMPIAR-10296 (Title: Single particle cryo-EM dataset of the triskelion hub subparticle extraction from clathrin cagesData size: 88.4 Data #1: Hub subparticles of the 28 mini coat [picked particles - multiframe - unprocessed] Data #2: Hub subparticles of the 32 sweet potato [picked particles - multiframe - unprocessed] Data #3: Hub subparticles of the 36 D6 barrel [picked particles - multiframe - unprocessed] Data #4: Hub subparticles of the 36 tennis ball [picked particles - multiframe - unprocessed] Data #5: Hub subparticles of the 37 big apple [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0122.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0122.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.705 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
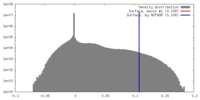
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0122_msk_1.map emd_0122_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
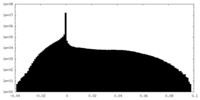
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Post processed map from resolution measurement.
| File | emd_0122_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post processed map from resolution measurement. | ||||||||||||
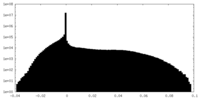
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Post processed map from resolution measurement.
| File | emd_0122_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post processed map from resolution measurement. | ||||||||||||
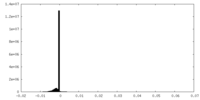
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Soft spherical masked (diameter 320A) applied to remove...
| File | emd_0122_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soft spherical masked (diameter 320A) applied to remove artifactual density prior to resolution measurement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Soft spherical masked (diameter 320A) applied to remove...
| File | emd_0122_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soft spherical masked (diameter 320A) applied to remove artifactual density prior to resolution measurement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Assembly of clathrin heavy and light chain into coat complexes
| Entire | Name: Assembly of clathrin heavy and light chain into coat complexes |
|---|---|
| Components |
|
-Supramolecule #1: Assembly of clathrin heavy and light chain into coat complexes
| Supramolecule | Name: Assembly of clathrin heavy and light chain into coat complexes type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 540 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.32 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.4 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER Details: 3 uL applied to a grid at room temperature and humidity. 3 second hand blot and plunge.. | |||||||||||||||
| Details | Clathrin coat complexes, end point assembly exhibiting architectural heterogeneity |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 3.0 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 82111 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.0032 µm / Nominal defocus min: 0.0014 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)