+ Open data
Open data
- Basic information
Basic information
| Entry | 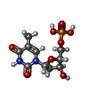 Database: PDB chemical components / ID: DT Database: PDB chemical components / ID: DT | ||
|---|---|---|---|
| Name | Name: | Comment | dTMP*YM | |
-Chemical information
| Composition |  | ||||
|---|---|---|---|---|---|
| Others | Type: DNA LINKING / PDB classification: ATOMN / One letter code: T / Three letter code: DT / Ideal coordinates details: OpenEye OEToolkits / Model coordinates PDB-ID: 214D / Replaces: T | ||||
| History |
| ||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  BindingDB / BindingDB /  Brenda Brenda                   / /  ChEBI / ChEBI /  DrugBank / DrugBank /  HMDB / HMDB /  KEGG_Ligand / KEGG_Ligand /  Metabolights / Metabolights /  NMRShiftDB / NMRShiftDB /  Nikkaji / Nikkaji /  PubChem / PubChem /  PubChem_TPharma / PubChem_TPharma /  SureChEMBL / SureChEMBL /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | [( | |
|---|
-PDB entries
Showing all 1 items

PDB-5cdm: 
2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA
 Movie
Movie Controller
Controller