+ Open data
Open data
- Basic information
Basic information
| Entry | 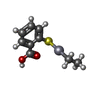 Database: PDB chemical components / ID: EMT Database: PDB chemical components / ID: EMT |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: EMT / Ideal coordinates details: Corina / Model coordinates PDB-ID: 1KDG / Replaces: T0M | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  BindingDB BindingDB                        / /  Brenda / Brenda /  ChEBI / ChEBI /  ChemicalBook / ChemicalBook /  CompTox / CompTox /  DrugBank / DrugBank /  Nikkaji / Nikkaji /  PubChem PubChem     / /  PubChem_TPharma / PubChem_TPharma /  SureChEMBL SureChEMBL     / /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | ( | |
|---|
-PDB entries
Showing all 5 items

PDB-1kdg: 
Crystal structure of the flavin domain of cellobiose dehydrogenase

PDB-1o9l: 
Succinate:Coenzyme-A Transferase (pig heart)

PDB-2ypd: 
Crystal structure of the Jumonji domain of human Jumonji domain containing 1C protein
PDB-3a0w: 
Catalytic domain of histidine kinase ThkA (TM1359) for MAD phasing (nucleotide free form 2, orthorombic)

PDB-4gsr: 
Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
 Movie
Movie Controller
Controller




