+ Open data
Open data
- Basic information
Basic information
| Entry | 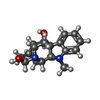 Database: PDB chemical components / ID: AJM Database: PDB chemical components / ID: AJM | ||
|---|---|---|---|
| Name | Name: | Comment | antiarrhythmic, alkaloid*YM | |
-Chemical information
| Composition |  | ||||
|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: AJM / Model coordinates PDB-ID: 1ZR8 | ||||
| History |
| ||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  PubChem / PubChem /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | ( |
|---|
-PDB entries
Showing all 4 items

PDB-1zr8: 
Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution

PDB-2que: 
Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution

PDB-3cbi: 
Crystal structure of the ternary complex of phospholipase A2 with ajmaline and anisic acid at 3.1 A resolution
PDB-3fg5: 
Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution
 Movie
Movie Controller
Controller



