+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9975 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
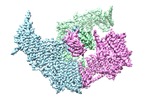
| Title | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Piezo / Mechanogating / Mechanotransduction channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmechanosensitive monoatomic cation channel activity / cuticular plate / detection of mechanical stimulus involved in sensory perception / mechanosensitive monoatomic ion channel activity / stereocilium / neuronal cell body membrane / monoatomic cation transport / response to mechanical stimulus / monoatomic cation channel activity / regulation of membrane potential / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Wang L / Zhou H / Zhang M / Liu W / Deng T / Zhao Q / Li Y / Lei J / Li X / Xiao B | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure and mechanogating of the mammalian tactile channel PIEZO2. Authors: Li Wang / Heng Zhou / Mingmin Zhang / Wenhao Liu / Tuan Deng / Qiancheng Zhao / Yiran Li / Jianlin Lei / Xueming Li / Bailong Xiao /  Abstract: PIEZO2 is a mechanosensitive cation channel that has a key role in sensing touch, tactile pain, breathing and blood pressure. Here we describe the cryo-electron microscopy structure of mouse PIEZO2, ...PIEZO2 is a mechanosensitive cation channel that has a key role in sensing touch, tactile pain, breathing and blood pressure. Here we describe the cryo-electron microscopy structure of mouse PIEZO2, which is a three-bladed, propeller-like trimer that comprises 114 transmembrane helices (38 per protomer). Transmembrane helices 1-36 (TM1-36) are folded into nine tandem units of four transmembrane helices each to form the unusual non-planar blades. The three blades are collectively curved into a nano-dome of 28-nm diameter and 10-nm depth, with an extracellular cap-like structure embedded in the centre and a 9-nm-long intracellular beam connecting to the central pore. TM38 and the C-terminal domain are surrounded by the anchor domain and TM37, and enclose the central pore with both transmembrane and cytoplasmic constriction sites. Structural comparison between PIEZO2 and its homologue PIEZO1 reveals that the transmembrane constriction site might act as a transmembrane gate that is controlled by the cap domain. Together, our studies provide insights into the structure and mechanogating mechanism of Piezo channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9975.map.gz emd_9975.map.gz | 9.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9975-v30.xml emd-9975-v30.xml emd-9975.xml emd-9975.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9975.png emd_9975.png | 255.2 KB | ||
| Filedesc metadata |  emd-9975.cif.gz emd-9975.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9975 http://ftp.pdbj.org/pub/emdb/structures/EMD-9975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9975 | HTTPS FTP |
-Related structure data
| Related structure data |  6kg7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9975.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9975.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.401 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : homo-trimer of Piezo2
| Entire | Name: homo-trimer of Piezo2 |
|---|---|
| Components |
|
-Supramolecule #1: homo-trimer of Piezo2
| Supramolecule | Name: homo-trimer of Piezo2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1 MDa |
-Macromolecule #1: Piezo-type mechanosensitive ion channel component 2
| Macromolecule | Name: Piezo-type mechanosensitive ion channel component 2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 325.984844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASEVVCGLI FRLLLPICLA VACAFRYNGL SFVYLIYLLL IPLFSEPTKA TMQGHTGRLL QSLCITSLSF LLLHIIFHIT LASLEAQHR ITPAYNCSTW EKTFRQIGFE SLKGADAGNG IRVFVPDIGM FIASLTIWLV CRTIVKKPDT EEIAQLNSEC E NEELAGGE ...String: MASEVVCGLI FRLLLPICLA VACAFRYNGL SFVYLIYLLL IPLFSEPTKA TMQGHTGRLL QSLCITSLSF LLLHIIFHIT LASLEAQHR ITPAYNCSTW EKTFRQIGFE SLKGADAGNG IRVFVPDIGM FIASLTIWLV CRTIVKKPDT EEIAQLNSEC E NEELAGGE KMDSEEALIY EEDLDGEEGM EGELEESTKL KILRRFASVA SKLKEFIGNM ITTAGKVVVT ILLGSSGMML PS LTSAVYF FVFLGLCTWW SWCRTFDPLL FGCLCVLLAI FTAGHLIGLY LYQFQFFQEA VPPNDYYARL FGIKSVIQTD CAS TWKIIV NPDLSWYHHA NPILLLVMYY TLATLIRIWL QEPLVQEEMA KEDEGALDCS SNQNTAERRR SLWYATQYPT DERK LLSMT QDDYKPSDGL LVTVNGNPVD YHTIHPSLPI ENGPAKTDLY TTPQYRWEPS EESSEKKEEE EDKREDSEGE GSQEE KRSV RMHAMVAVFQ FIMKQSYICA LIAMMAWSIT YHSWLTFVLL IWSCTLWMIR NRRKYAMISS PFMVVYANLL LVLQYI WSF ELPEIKKVPG FLEKKEPGEL ASKILFTITF WLLLRQHLTE QKALREKEAL LSEVKIGSQE LEEKEDEELQ DVQVEGE PT EKEEEEEEEI KEERHEVKKE EEEEVEEDDD QDIMKVLGNL VVALFIKYWI YVCGGMFFFV SFEGKIVMYK IIYMVLFL F CVALYQVHYE WWRKILKYFW MSVVIYTMLV LIFIYTYQFE NFPGLWQNMT GLKKEKLEDL GLKQFTVAEL FTRIFIPTS FLLVCILHLH YFHDRFLELT DLKSIPSKED NTIYSHAKVN GRVYLIINRL AHPEGSLPDL AIMNMTASLD KPEVQKLAES GEERPEECV KKTEKGEAGK DSDESEEEED EEEESEEEES SDLRNKWHLV IDRLTVLFLK FLEYFHKLQV FMWWILELHI I KIVSSYII WVTVKEVSLF NYVFLISWAF ALPYAKLRRA ASSVCTVWTC VIIVCKMLYQ LQTIKPENFS VNCSLPNENQ TN IPLHELN KSLLYSAPVD PTEWVGLRKS SPLLVYLRNN LLMLAILAFE VTVYRHQEYY RGRNNLTAPV SKTIFHDITR LHL DDGLIN CAKYFVNYFF YKFGLETCFL MSVNVIGQRM DFYAMIHACW LIGVLYRRRR KAIAEVWPKY CCFLACIITF QYFV CIGIP PAPCRDYPWR FKGAYFNDNI IKWLYFPDFI VRPNPVFLVY DFMLLLCASL QRQIFEDENK AAVRIMAGDN VEICM NLDA ASFSQHNPVP DFIHCRSYLD MSKVIIFSYL FWFVLTIIFI TGTTRISIFC MGYLVACFYF LLFGGDLLLK PIKSIL RYW DWLIAYNVFV ITMKNILSIG ACGYIGALVR NSCWLIQAFS LACTVKGYQM PEDDSRCKLP SGEAGIIWDS ICFAFLL LQ RRVFMSYYFL HVVADIKASQ ILASRGAELF QATIVKAVKA RIEEEKKSMD QLKRQMDRIK ARQQKYKKGK ERMLSLTQ E SGEGQDIQKV SEEDDEREAD KQKAKGKKKQ WWRPWVDHAS MVRSGDYYLF ETDSEEEEEE ELKKEDEEPP RKSAFQFVY QAWITDPKTA LRQRRKEKKK LAREEQKERR KGSGDGPVEW EDREDEPVKK KSDGPDNIIK RIFNILKFTW VLFLATVDSF TTWLNSISR EHIDISTVLR IERCMLTREI KKGNVPTRES IHMYYQNHIM NLSRESGLDT IDEHSGAGSR AQAAHRMDSL D SRDSISSC YTEATLLISR QSTLDDLDGQ DPVPKTSERA RPRLRKMFSL DMSSSSADSG SVASSEPTQC TMLYSRQGTT ET IEEVEAE AEEEVVEGLE PELHDAEEKE YAAEYEAGVE EISLTPDEEL PQFSTDDCEA PPSYSKAVSF EHLSFASQDD SGA KNHMVV SPDDSRTDKL ESSILPPLTH ELTASDLLMS KMFHDDELEE SEKFYVDQPR FLLLFYAMYN TLVARSEMVC YFVI ILNHM TSASIITLLL PILIFLWAML SVPRPSRRFW MMAIVYTEVA IVVKYFFQFG FFPWNKDLEI YKERPYFPPN IIGVE KKEG YVLYDLIQLL ALFFHRSILK CHGLWDEDDI VDSNTDKEGS DDELSLDQGR RGSSDSLKSI NLAASVESVH VTFPEQ PAA IRRKRSCSSS QISPRSSFSS NRSKRGSTST RNSSQKGSSV LSLKQKSKRE LYMEKLQEHL IKAKAFTIKK TLQIYVP IR QFFYDLIHPD YSAVTDVYVL MFLADTVDFI IIVFGFWAFG KHSAAADITS SLSEDQVPGP FLVMVLIQFG TMVVDRAL Y LRKTVLGKVI FQVILVFGIH FWMFFILPGV TERKFSQNLV AQLWYFVKCV YFGLSAYQIR CGYPTRVLGN FLTKSYNYV NLFLFQGFRL VPFLTELRAV MDWVWTDTTL SLSSWICVED IYAHIFILKC WRESEKRYPQ PRGQKKKKAV KYGMGGMIIV LLICIVWFP LLFMSLIKSV AGVINQPLDV SVTITLGGYQ PIFTMSAQQS QLKVMDNSKY NEFLKSFGPN SGAMQFLENY E REDVTVAE LEGNSNSLWT ISPPSKQKMI QELTDPNSCF SVVFSWSIQR NMTLGAKAEI ATDKLSFPLA VATRNSIAKM IA GNDTESS NTPVTIEKIY PYYVKAPSDS NSKPIKQLLS ENNFMNITII LFRDNVTKSN SEWWVLNLTG SRIFNQGSQA LEL VVFNDK VSPPSLGFLA GYGIMGLYAS VVLVIGKFVR EFFSGISHSI MFEELPNVDR ILKLCTDIFL VRETGELELE EDLY AKLIF LYRSPETMIK WTREKTN UniProtKB: Piezo-type mechanosensitive ion channel component 2 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 2721959 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6kg7: |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)






















