+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9796 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
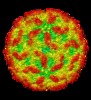
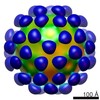
| Title | AAV5 in complex with AAVR | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | adeno-associated virus / AAV5 / receptor / AAVR / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationproacrosomal vesicle fusion / response to auditory stimulus / flagellated sperm motility / T=1 icosahedral viral capsid / receptor-mediated endocytosis of virus by host cell / trans-Golgi network / neuron migration / cytoplasmic vesicle / Golgi membrane / nucleolus ...proacrosomal vesicle fusion / response to auditory stimulus / flagellated sperm motility / T=1 icosahedral viral capsid / receptor-mediated endocytosis of virus by host cell / trans-Golgi network / neuron migration / cytoplasmic vesicle / Golgi membrane / nucleolus / structural molecule activity / Golgi apparatus / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Adeno-associated virus - 5 / Adeno-associated virus - 5 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.18 Å | |||||||||
 Authors Authors | Lou Z / Zhang R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Divergent engagements between adeno-associated viruses with their cellular receptor AAVR. Authors: Ran Zhang / Guangxue Xu / Lin Cao / Zixian Sun / Yong He / Mengtian Cui / Yuna Sun / Shentao Li / Huapeng Li / Lan Qin / Mingxu Hu / Zhengjia Yuan / Zipei Rao / Wei Ding / Zihe Rao / Zhiyong Lou /  Abstract: Adeno-associated virus (AAV) receptor (AAVR) is an essential receptor for the entry of multiple AAV serotypes with divergent rules; however, the mechanism remains unclear. Here, we determine the ...Adeno-associated virus (AAV) receptor (AAVR) is an essential receptor for the entry of multiple AAV serotypes with divergent rules; however, the mechanism remains unclear. Here, we determine the structures of the AAV1-AAVR and AAV5-AAVR complexes, revealing the molecular details by which PKD1 recognizes AAV5 and PKD2 is solely engaged with AAV1. PKD2 lies on the plateau region of the AAV1 capsid. However, the AAV5-AAVR interface is strikingly different, in which PKD1 is bound at the opposite side of the spike of the AAV5 capsid than the PKD2-interacting region of AAV1. Residues in strands F/G and the CD loop of PKD1 interact directly with AAV5, whereas residues in strands B/C/E and the BC loop of PKD2 make contact with AAV1. These findings further the understanding of the distinct mechanisms by which AAVR recognizes various AAV serotypes and provide an example of a single receptor engaging multiple viral serotypes with divergent rules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9796.map.gz emd_9796.map.gz | 64.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9796-v30.xml emd-9796-v30.xml emd-9796.xml emd-9796.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9796.png emd_9796.png | 241.2 KB | ||
| Filedesc metadata |  emd-9796.cif.gz emd-9796.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9796 http://ftp.pdbj.org/pub/emdb/structures/EMD-9796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9796 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9796 | HTTPS FTP |
-Validation report
| Summary document |  emd_9796_validation.pdf.gz emd_9796_validation.pdf.gz | 664.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9796_full_validation.pdf.gz emd_9796_full_validation.pdf.gz | 664.2 KB | Display | |
| Data in XML |  emd_9796_validation.xml.gz emd_9796_validation.xml.gz | 7.2 KB | Display | |
| Data in CIF |  emd_9796_validation.cif.gz emd_9796_validation.cif.gz | 8.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9796 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9796 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9796 | HTTPS FTP |
-Related structure data
| Related structure data |  6jcsMC  9794C  9795C  9797C  6jcqC  6jcrC  6jctC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9796.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9796.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
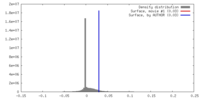
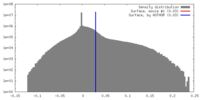
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus - 5
| Entire | Name:  Adeno-associated virus - 5 Adeno-associated virus - 5 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus - 5
| Supramolecule | Name: Adeno-associated virus - 5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: capsid protein VP1
| Supramolecule | Name: capsid protein VP1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 5 Adeno-associated virus - 5 |
-Supramolecule #3: PKD1
| Supramolecule | Name: PKD1 / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 5 Adeno-associated virus - 5 |
| Molecular weight | Theoretical: 58.213078 KDa |
| Sequence | String: DGVGNASGDW HCDSTWMGDR VVTKSTRTWV LPSYNNHQYR EIKSGSVDGS NANAYFGYST PWGYFDFNRF HSHWSPRDWQ RLINNYWGF RPRSLRVKIF NIQVKEVTVQ DSTTTIANNL TSTVQVFTDD DYQLPYVVGN GTEGCLPAFP PQVFTLPQYG Y ATLNRDNT ...String: DGVGNASGDW HCDSTWMGDR VVTKSTRTWV LPSYNNHQYR EIKSGSVDGS NANAYFGYST PWGYFDFNRF HSHWSPRDWQ RLINNYWGF RPRSLRVKIF NIQVKEVTVQ DSTTTIANNL TSTVQVFTDD DYQLPYVVGN GTEGCLPAFP PQVFTLPQYG Y ATLNRDNT ENPTERSSFF CLEYFPSKML RTGNNFEFTY NFEEVPFHSS FAPSQNLFKL ANPLVDQYLY RFVSTNNTGG VQ FNKNLAG RYANTYKNWF PGPMGRTQGW NLGSGVNRAS VSAFATTNRM ELEGASYQVP PQPNGMTNNL QGSNTYALEN TMI FNSQPA NPGTTATYLE GNMLITSESE TQPVNRVAYN VGGQMATNNQ SSTTAPATGT YNLQEIVPGS VWMERDVYLQ GPIW AKIPE TGAHFHPSPA MGGFGLKHPP PMMLIKNTPV PGNITSFSDV PVSSFITQYS TGQVTVEMEW ELKKENSKRW NPEIQ YTNN YNDPQFVDFA PDSTGEYRTT RPIGTRYLTR PL UniProtKB: Capsid protein |
-Macromolecule #2: Dyslexia-associated protein KIAA0319-like protein
| Macromolecule | Name: Dyslexia-associated protein KIAA0319-like protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.911322 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VIKELVVSAG ESVQITLPKN EVQLNAYVLQ EPPKGETYTY DWQLITHPRD YSGEMEGKHS QILKLSKLTP GLYEFKVIVE GQNAHGEGY VNVTVKPE UniProtKB: Dyslexia-associated protein KIAA0319-like protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)