[English] 日本語
 Yorodumi
Yorodumi- EMDB-9696: Cryo-EM structure of multidrug efflux pump MexAB-OprM (60 degree ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9696 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of multidrug efflux pump MexAB-OprM (60 degree state) | ||||||||||||
 Map data Map data | Sharpened with B-factor sharpening. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Multidrug resistance / efflux / complex / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationefflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / transmembrane transporter activity / cell outer membrane / protein homooligomerization / transmembrane transport / response to antibiotic / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | ||||||||||||
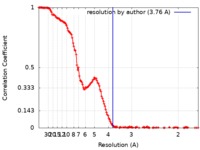
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | ||||||||||||
 Authors Authors | Tsutsumi K / Yonehara R | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism. Authors: Kenta Tsutsumi / Ryo Yonehara / Etsuko Ishizaka-Ikeda / Naoyuki Miyazaki / Shintaro Maeda / Kenji Iwasaki / Atsushi Nakagawa / Eiki Yamashita /   Abstract: In Pseudomonas aeruginosa, MexAB-OprM plays a central role in multidrug resistance by ejecting various drug compounds, which is one of the causes of serious nosocomial infections. Although the ...In Pseudomonas aeruginosa, MexAB-OprM plays a central role in multidrug resistance by ejecting various drug compounds, which is one of the causes of serious nosocomial infections. Although the structures of the components of MexAB-OprM have been solved individually by X-ray crystallography, no structural information for fully assembled pumps from P. aeruginosa were previously available. In this study, we present the structure of wild-type MexAB-OprM in the presence or absence of drugs at near-atomic resolution. The structure reveals that OprM does not interact with MexB directly, and that it opens its periplasmic gate by forming a complex. Furthermore, we confirm the residues essential for complex formation and observed a movement of the drug entrance gate. Based on these results, we propose mechanisms for complex formation and drug efflux. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9696.map.gz emd_9696.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9696-v30.xml emd-9696-v30.xml emd-9696.xml emd-9696.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9696_fsc.xml emd_9696_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_9696.png emd_9696.png | 80.8 KB | ||
| Filedesc metadata |  emd-9696.cif.gz emd-9696.cif.gz | 7.1 KB | ||
| Others |  emd_9696_half_map_1.map.gz emd_9696_half_map_1.map.gz emd_9696_half_map_2.map.gz emd_9696_half_map_2.map.gz | 193.8 MB 193.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9696 http://ftp.pdbj.org/pub/emdb/structures/EMD-9696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9696 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9696 | HTTPS FTP |
-Validation report
| Summary document |  emd_9696_validation.pdf.gz emd_9696_validation.pdf.gz | 736.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9696_full_validation.pdf.gz emd_9696_full_validation.pdf.gz | 735.9 KB | Display | |
| Data in XML |  emd_9696_validation.xml.gz emd_9696_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_9696_validation.cif.gz emd_9696_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9696 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9696 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9696 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9696 | HTTPS FTP |
-Related structure data
| Related structure data |  6iolMC  9695C  6iokC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10292 (Title: Cryo-EM structure of multidrug efflux pump MexAB-OprM EMPIAR-10292 (Title: Cryo-EM structure of multidrug efflux pump MexAB-OprMData size: 8.5 TB Data #1: Raw multiframe micrographs [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9696.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9696.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened with B-factor sharpening. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.875 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_9696_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_9696_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : membrane protein complex
| Entire | Name: membrane protein complex |
|---|---|
| Components |
|
-Supramolecule #1: membrane protein complex
| Supramolecule | Name: membrane protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 730 KDa |
-Macromolecule #1: Multidrug resistance protein MexA
| Macromolecule | Name: Multidrug resistance protein MexA / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.789695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGKSEAPP PAQTPEVGIV TLEAQTVTLN TELPGRTNAF RIAEVRPQVN GIILKRLFKE GSDVKAGQQL YQIDPATYEA DYQSAQANL ASTQEQAQRY KLLVADQAVS KQQYADANAA YLQSKAAVEQ ARINLRYTKV LSPISGRIGR SAVTEGALVT N GQANAMAT ...String: GAMGKSEAPP PAQTPEVGIV TLEAQTVTLN TELPGRTNAF RIAEVRPQVN GIILKRLFKE GSDVKAGQQL YQIDPATYEA DYQSAQANL ASTQEQAQRY KLLVADQAVS KQQYADANAA YLQSKAAVEQ ARINLRYTKV LSPISGRIGR SAVTEGALVT N GQANAMAT VQQLDPIYVD VTQPSTALLR LRRELASGQL ERAGDNAAKV SLKLEDGSQY PLEGRLEFSE VSVDEGTGSV TI RAVFPNP NNELLPGMFV HAQLQEGVKQ KAILAPQQGV TRDLKGQATA LVVNAQNKVE LRVIKADRVI GDKWLVTEGL NAG DKIITE GLQFVQPGVE VKTVPAKNVA SAQKADAAPA KTDSKG UniProtKB: Multidrug resistance protein MexA |
-Macromolecule #2: Outer membrane protein OprM
| Macromolecule | Name: Outer membrane protein OprM / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 51.737605 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSLIPDYQRP EAPVAAAYPQ GQAYGQNTGA AAVPAADIGW REFFRDPQLQ QLIGVALENN RDLRVAALNV EAFRAQYRIQ RADLFPRIG VDGSGTRQRL PGDLSTTGSP AISSQYGVTL GTTAWELDLF GRLRSLRDQA LEQYLATEQA QRSAQTTLVA S VATAYLTL ...String: CSLIPDYQRP EAPVAAAYPQ GQAYGQNTGA AAVPAADIGW REFFRDPQLQ QLIGVALENN RDLRVAALNV EAFRAQYRIQ RADLFPRIG VDGSGTRQRL PGDLSTTGSP AISSQYGVTL GTTAWELDLF GRLRSLRDQA LEQYLATEQA QRSAQTTLVA S VATAYLTL KADQAQLQLT KDTLGTYQKS FDLTQRSYDV GVASALDLRQ AQTAVEGARA TLAQYTRLVA QDQNALVLLL GS GIPANLP QGLGLDQTLL TEVPAGLPSD LLQRRPDILE AEHQLMAANA SIGAARAAFF PSISLTANAG TMSRQLSGLF DAG SGSWLF QPSINLPIFT AGSLRASLDY AKIQKDINVA QYEKAIQTAF QEVADGLAAR GTFTEQLQAQ RDLVKASDEY YQLA DKRYR TGVDNYLTLL DAQRSLFTAQ QQLITDRLNQ LTSEVNLYKA LGGGWNQQTV TQQQTAKKED PQAHHHHHH UniProtKB: Outer membrane protein OprM |
-Macromolecule #3: Multidrug resistance protein MexB
| Macromolecule | Name: Multidrug resistance protein MexB / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 Pseudomonas aeruginosa PAO1 (bacteria) / Strain: PAO1 |
| Molecular weight | Theoretical: 113.948273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKFFIDRPI FAWVIALVIM LAGGLSILSL PVNQYPAIAP PAIAVQVSYP GASAETVQDT VVQVIEQQMN GIDNLRYISS ESNSDGSMT ITVTFEQGTD PDIAQVQVQN KLQLATPLLP QEVQRQGIRV TKAVKNFLMV VGVVSTDGSM TKEDLSNYIV S NIQDPLSR ...String: MSKFFIDRPI FAWVIALVIM LAGGLSILSL PVNQYPAIAP PAIAVQVSYP GASAETVQDT VVQVIEQQMN GIDNLRYISS ESNSDGSMT ITVTFEQGTD PDIAQVQVQN KLQLATPLLP QEVQRQGIRV TKAVKNFLMV VGVVSTDGSM TKEDLSNYIV S NIQDPLSR TKGVGDFQVF GSQYSMRIWL DPAKLNSYQL TPGDVSSAIQ AQNVQISSGQ LGGLPAVKGQ QLNATIIGKT RL QTAEQFE NILLKVNPDG SQVRLKDVAD VGLGGQDYSI NAQFNGSPAS GIAIKLATGA NALDTAKAIR QTIANLEPFM PQG MKVVYP YDTTPVVSAS IHEVVKTLGE AILLVFLVMY LFLQNFRATL IPTIAVPVVL LGTFGVLAAF GFSINTLTMF GMVL AIGLL VDDAIVVVEN VERVMAEEGL SPREAARKSM GQIQGALVGI AMVLSAVFLP MAFFGGSTGV IYRQFSITIV SAMAL SVIV ALILTPALCA TMLKPIEKGD HGEHKGGFFG WFNRMFLSTT HGYERGVASI LKHRAPYLLI YVVIVAGMIW MFTRIP TAF LPDEDQGVLF AQVQTPPGSS AERTQVVVDS MREYLLEKES SSVSSVFTVT GFNFAGRGQS SGMAFIMLKP WEERPGG EN SVFELAKRAQ MHFFSFKDAM VFAFAPPSVL ELGNATGFDL FLQDQAGVGH EVLLQARNKF LMLAAQNPAL QRVRPNGM S DEPQYKLEID DEKASALGVS LADINSTVSI AWGSSYVNDF IDRGRVKRVY LQGRPDARMN PDDLSKWYVR NDKGEMVPF NAFATGKWEY GSPKLERYNG VPAMEILGEP APGLSSGDAM AAVEEIVKQL PKGVGYSWTG LSYEERLSGS QAPALYALSL LVVFLCLAA LYESWSIPFS VMLVVPLGVI GALLATSMRG LSNDVFFQVG LLTTIGLSAK NAILIVEFAK ELHEQGKGIV E AAIEACRM RLRPIVMTSL AFILGVVPLA ISTGAGSGSQ HAIGTGVIGG MVTATVLAIF WVPLFYVAVS TLFKDEASKQ QA SVEKGQL EHHHHHH UniProtKB: Multidrug resistance protein MexB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 50.0 mM / Component - Formula: C8H18N2O4S / Component - Name: HEPES |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.0 K / Max: 95.0 K |
| Specialist optics | Spherical aberration corrector: CEOS Cs-corrector |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4080 pixel / Digitization - Dimensions - Height: 4080 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Number real images: 8722 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.1 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)