[English] 日本語
 Yorodumi
Yorodumi- EMDB-9279: CryoEM structure of Chimeric Eastern Equine Encephalitis Virus wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9279 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-69 Antibody | |||||||||||||||
 Map data Map data | EEEV-69 Fab full map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Alphavirus / EEEV / Eastern Equine Encephalitis Virus / Sindbis / Fab / VIRUS-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host gene expression / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host gene expression / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell / host cell plasma membrane / structural molecule activity / virion membrane / proteolysis / RNA binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   Eastern equine encephalitis virus / Eastern equine encephalitis virus /  | |||||||||||||||
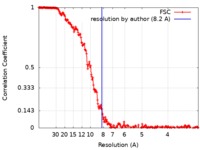
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | |||||||||||||||
 Authors Authors | Hasan SS / Sun C | |||||||||||||||
| Funding support |  United States, United States,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2018 Journal: Cell Rep / Year: 2018Title: Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization. Authors: S Saif Hasan / Chengqun Sun / Arthur S Kim / Yasunori Watanabe / Chun-Liang Chen / Thomas Klose / Geeta Buda / Max Crispin / Michael S Diamond / William B Klimstra / Michael G Rossmann /   Abstract: Alphaviruses are enveloped pathogens that cause arthritis and encephalitis. Here, we report a 4.4-Å cryoelectron microscopy (cryo-EM) structure of eastern equine encephalitis virus (EEEV), an ...Alphaviruses are enveloped pathogens that cause arthritis and encephalitis. Here, we report a 4.4-Å cryoelectron microscopy (cryo-EM) structure of eastern equine encephalitis virus (EEEV), an alphavirus that causes fatal encephalitis in humans. Our analysis provides insights into viral entry into host cells. The envelope protein E2 showed a binding site for the cellular attachment factor heparan sulfate. The presence of a cryptic E2 glycan suggests how EEEV escapes surveillance by lectin-expressing myeloid lineage cells, which are sentinels of the immune system. A mechanism for nucleocapsid core release and disassembly upon viral entry was inferred based on pH changes and capsid dissociation from envelope proteins. The EEEV capsid structure showed a viral RNA genome binding site adjacent to a ribosome binding site for viral genome translation following genome release. Using five Fab-EEEV complexes derived from neutralizing antibodies, our investigation provides insights into EEEV host cell interactions and protective epitopes relevant to vaccine design. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9279.map.gz emd_9279.map.gz | 535 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9279-v30.xml emd-9279-v30.xml emd-9279.xml emd-9279.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9279_fsc.xml emd_9279_fsc.xml | 23.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_9279.png emd_9279.png | 222.8 KB | ||
| Filedesc metadata |  emd-9279.cif.gz emd-9279.cif.gz | 6.8 KB | ||
| Others |  emd_9279_half_map_1.map.gz emd_9279_half_map_1.map.gz emd_9279_half_map_2.map.gz emd_9279_half_map_2.map.gz | 369.4 MB 369.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9279 http://ftp.pdbj.org/pub/emdb/structures/EMD-9279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9279 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9279 | HTTPS FTP |
-Validation report
| Summary document |  emd_9279_validation.pdf.gz emd_9279_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9279_full_validation.pdf.gz emd_9279_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_9279_validation.xml.gz emd_9279_validation.xml.gz | 28.1 KB | Display | |
| Data in CIF |  emd_9279_validation.cif.gz emd_9279_validation.cif.gz | 37.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9279 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9279 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9279 | HTTPS FTP |
-Related structure data
| Related structure data |  6mwxMC  9249C  9274C  9275C  9278C  9280C  9281C  6muiC  6mw9C  6mwcC  6mwvC  6mx4C  6mx7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9279.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9279.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV-69 Fab full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: EEEV-69 Fab half map #1
| File | emd_9279_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV-69 Fab half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EEEV Fab69 half map #2
| File | emd_9279_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV Fab69 half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern equine encephalitis virus / EEEV-69 Fab complex
| Entire | Name: Eastern equine encephalitis virus / EEEV-69 Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Eastern equine encephalitis virus / EEEV-69 Fab complex
| Supramolecule | Name: Eastern equine encephalitis virus / EEEV-69 Fab complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purified from infected BHK-15 cells |
|---|
-Supramolecule #2: Eastern equine encephalitis virus
| Supramolecule | Name: Eastern equine encephalitis virus / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
-Supramolecule #3: EEEV-69 Fab
| Supramolecule | Name: EEEV-69 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: E1
| Macromolecule | Name: E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.938141 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FTGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS ...String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FTGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS SAWSPFDNKV VVYGHEVYNY DFPEYGTGKA GSFGDLQSRT STSNDLYANT NLKLQRPQAG IVHTPFTQAP SG FERWKRD KGAPLNDVAP FGCSIALEPL RAENCAVGSI PISIDIPDAA FTRISETPTV SDLECKITEC TYASDFGGIA TVA YKSSKA GNCPIHSPSG VAVIKENDVT LAESGSFTFH FSTANIHPAF KLQVCTSAVT CKGDCKPPKD HIVDYPAQHT ESFT SAISA TAWSWLKVLV GGTSAFIVLG LIATAVVALV LFFHRH UniProtKB: Structural polyprotein |
-Macromolecule #2: E2
| Macromolecule | Name: E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.046953 KDa |
| Recombinant expression | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ ...String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ PGLVADHSLL SIHSAKVKIT VPSGAQVKYY CKCPDVREGI TSSDHTTTCT DVKQCRAYLI DNKKWVYNSG RL PRGEGDT FKGKLHVPFV PVKAKCIATL APEPLVEHKH RTLILHLHPD HPTLLTTRSL GSDANPTRQW IERPTTVNFT VTG EGLEYT WGNHPPKRVW AQESGEGNPH GWPHEVVVYY YNRYPLTTII GLCTCVAIIM VSCVTSVWLL CRTRNLCITP YKLA PNAQV PILLALLCCI KPTRA UniProtKB: Structural polyprotein |
-Macromolecule #3: EEEV-69 antibody heavy chain
| Macromolecule | Name: EEEV-69 antibody heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.743719 KDa |
| Sequence | String: QIQLVQSGRE VKNPGETVKI SCKASGYTFT EYPMLWVKQA PGKGFRWMGL IYTNTGEPTY AEEFKGRFVF SLEISASTAY LQINNLTNE DTATYFCVRD YFISLDYWGQ GTTLTVSSAK TTAPSVYPLA PVCGGTTGSS VTLGCLVKGY FPEPVTLTWN S GSLSSGVH ...String: QIQLVQSGRE VKNPGETVKI SCKASGYTFT EYPMLWVKQA PGKGFRWMGL IYTNTGEPTY AEEFKGRFVF SLEISASTAY LQINNLTNE DTATYFCVRD YFISLDYWGQ GTTLTVSSAK TTAPSVYPLA PVCGGTTGSS VTLGCLVKGY FPEPVTLTWN S GSLSSGVH TFPALLQSGL YTLSSSVTVT SNTWPSQTIT CNVAHPASST KVDKKIESRR |
-Macromolecule #4: EEEV-69 antibody light chain
| Macromolecule | Name: EEEV-69 antibody light chain / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.177713 KDa |
| Sequence | String: QAVVTQESAL TTSPGETVTL TCRSNIGAVT SSNCANWVQE KPDHFFTGLI GDTNNRRSGV PARFSGSLIG DKAALTITGA QTEDEAIYF CALWYNNLWV FGGGTKLTVL GQPKSSPSVT LFPPSSEELE TNKATLVCTI TDFYPGVVTV DWKVDGTPVT Q GMETTQPS ...String: QAVVTQESAL TTSPGETVTL TCRSNIGAVT SSNCANWVQE KPDHFFTGLI GDTNNRRSGV PARFSGSLIG DKAALTITGA QTEDEAIYF CALWYNNLWV FGGGTKLTVL GQPKSSPSVT LFPPSSEELE TNKATLVCTI TDFYPGVVTV DWKVDGTPVT Q GMETTQPS KQSNNKYMAS SYLTLTARAW ERHSSYSCQV THEGHTVEKS LSRADC UniProtKB: Anti-human Langerin 2G3 lambda chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: HELIUM / Chamber humidity: 80 % / Chamber temperature: 295 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6mwx: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)