[English] 日本語
 Yorodumi
Yorodumi- EMDB-9107: A unique supramolecular organization of photosystem I in the moss... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9107 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A unique supramolecular organization of photosystem I in the moss Physcomitrella patens | |||||||||
 Map data Map data | Large Physcomitrella patens PSI-LHCI supercomplex map from cryo-EM | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | photosynthesis / complex / photosystem I / light harvesting complex / Physcomitrella | |||||||||
| Biological species |  Physcomitrella patens (plant) Physcomitrella patens (plant) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.6 Å | |||||||||
 Authors Authors | Iwai M / Grob P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2018 Journal: Nat Plants / Year: 2018Title: A unique supramolecular organization of photosystem I in the moss Physcomitrella patens. Authors: Masakazu Iwai / Patricia Grob / Anthony T Iavarone / Eva Nogales / Krishna K Niyogi /  Abstract: The photosynthesis machinery in chloroplast thylakoid membranes is comprised of multiple protein complexes and supercomplexes. Here, we show a novel supramolecular organization of photosystem I (PSI) ...The photosynthesis machinery in chloroplast thylakoid membranes is comprised of multiple protein complexes and supercomplexes. Here, we show a novel supramolecular organization of photosystem I (PSI) in the moss Physcomitrella patens by single-particle cryo-electron microscopy. The moss-specific light-harvesting complex (LHC) protein Lhcb9 is involved in this PSI supercomplex, which has been shown to have a molecular density similar to that of the green alga Chlamydomonas reinhardtii. Our results show that the structural organization is unexpectedly different-two rows of the LHCI belt exist as in C. reinhardtii, but the outer one is shifted toward the PsaK side. Furthermore, one trimeric LHC protein and one monomeric LHC protein position alongside PsaL/K, filling the gap between these subunits and the outer LHCI belt. We provide evidence showing that Lhcb9 is a key factor, acting as a linkage between the PSI core and the outer LHCI belt to form the unique supramolecular organization of the PSI supercomplex in P. patens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9107.map.gz emd_9107.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9107-v30.xml emd-9107-v30.xml emd-9107.xml emd-9107.xml | 55.4 KB 55.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9107.png emd_9107.png | 294.5 KB | ||
| Filedesc metadata |  emd-9107.cif.gz emd-9107.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9107 http://ftp.pdbj.org/pub/emdb/structures/EMD-9107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9107 | HTTPS FTP |
-Related structure data
| Related structure data |  6memMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9107.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9107.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Large Physcomitrella patens PSI-LHCI supercomplex map from cryo-EM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
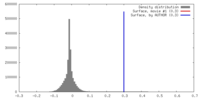
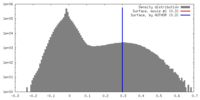
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Large PSI-LHCI supercomplex
+Supramolecule #1: Large PSI-LHCI supercomplex
+Macromolecule #1: Chlorophyll A/B binding protein 1
+Macromolecule #2: Chlorophyll A/B binding protein 5
+Macromolecule #3: Chlorophyll A/B binding protein 3
+Macromolecule #4: Chlorophyll A/B binding protein 7
+Macromolecule #5: Chlorophyll A/B binding protein 4
+Macromolecule #6: Chlorophyll A/B binding protein 9
+Macromolecule #7: Chlorophyll A/B binding protein 8
+Macromolecule #8: Chlorophyll A/B binding protein 2
+Macromolecule #9: Chlorophyll A/B binding protein 6
+Macromolecule #10: PsaA
+Macromolecule #11: Chlorophyll A/B binding protein 10
+Macromolecule #12: PsaB
+Macromolecule #13: Chlorophyll A/B binding protein 11
+Macromolecule #14: PsaC
+Macromolecule #15: Chlorophyll A/B binding protein 12
+Macromolecule #16: PsaD
+Macromolecule #17: PsaE
+Macromolecule #18: PsaF
+Macromolecule #19: PsaG
+Macromolecule #20: PsaH
+Macromolecule #21: PsaI
+Macromolecule #22: PsaJ
+Macromolecule #23: PsaK
+Macromolecule #24: PsaL
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Lights off, blot 4.5 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 3 / Number real images: 361 / Average exposure time: 0.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: -4.2 µm / Calibrated defocus min: -1.9000000000000001 µm / Calibrated magnification: 107140 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: -1.8 µm / Nominal defocus min: -2.5 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6mem: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)