[English] 日本語
 Yorodumi
Yorodumi- PDB-8cyi: Cryo-EM structures and computational analysis for enhanced potenc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8cyi | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ONCOPROTEIN/Transferase / PRMT5 / protein arginine methyl transferase / MTA-inhibitor synergy / cryo-EM structure-based drug design / computational analysis / catalytic mechanism / drug discovery / docking analysis / ONCOPROTEIN / ONCOPROTEIN-Transferase complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway / peptidyl-arginine N-methylation / oocyte axis specification / type II protein arginine methyltransferase / protein-arginine omega-N symmetric methyltransferase activity / peptidyl-arginine methylation / Golgi ribbon formation / negative regulation of epithelial cell proliferation involved in prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / histone H4R3 methyltransferase activity ...positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway / peptidyl-arginine N-methylation / oocyte axis specification / type II protein arginine methyltransferase / protein-arginine omega-N symmetric methyltransferase activity / peptidyl-arginine methylation / Golgi ribbon formation / negative regulation of epithelial cell proliferation involved in prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / histone H4R3 methyltransferase activity / : / epithelial cell proliferation involved in prostate gland development / protein-arginine N-methyltransferase activity / methylosome / positive regulation of mRNA splicing, via spliceosome / methyl-CpG binding / endothelial cell activation / histone H3 methyltransferase activity / histone methyltransferase activity / regulation of mitotic nuclear division / negative regulation of gene expression via chromosomal CpG island methylation / Cul4B-RING E3 ubiquitin ligase complex / E-box binding / histone methyltransferase complex / positive regulation of oligodendrocyte differentiation / negative regulation of cell differentiation / spliceosomal snRNP assembly / ribonucleoprotein complex binding / regulation of ERK1 and ERK2 cascade / ubiquitin-like ligase-substrate adaptor activity / liver regeneration / regulation of signal transduction by p53 class mediator / methyltransferase activity / circadian regulation of gene expression / DNA-templated transcription termination / Regulation of TP53 Activity through Methylation / RMTs methylate histone arginines / protein polyubiquitination / p53 binding / transcription corepressor activity / snRNP Assembly / ubiquitin-dependent protein catabolic process / transcription coactivator activity / chromatin remodeling / protein heterodimerization activity / positive regulation of cell population proliferation / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / Golgi apparatus / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.14 Å | ||||||
 Authors Authors | Yadav, G.P. / Wei, Z. / Xiaozhi, Y. / Chenglong, L. / Jiang, Q. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
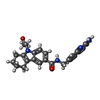
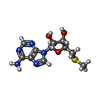
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency. Authors: Wei Zhou / Gaya P Yadav / Xiaozhi Yang / Feng Qin / Chenglong Li / Qiu-Xing Jiang /  Abstract: Projected potential of 2.5-4.0 Å cryo-EM structures for structure-based drug design is not well realized yet. Here we show that a 3.1 Å structure of PRMT5 is suitable for selecting computed ...Projected potential of 2.5-4.0 Å cryo-EM structures for structure-based drug design is not well realized yet. Here we show that a 3.1 Å structure of PRMT5 is suitable for selecting computed poses of a chemical inhibitor and its analogs for enhanced potency. PRMT5, an oncogenic target for various cancer types, has many inhibitors manifesting little cooperativity with MTA, a co-factor analog accumulated in MTAP-/- cells. To achieve MTA-synergic inhibition, a pharmacophore from virtual screen leads to a specific inhibitor (11-2 F). Cryo-EM structures of 11-2 F / MTA-bound human PRMT5/MEP50 complex and its apo form resolved at 3.1 and 3.2 Å respectively show that 11-2 F in the catalytic pocket shifts the cofactor-binding pocket away by ~2.0 Å, contributing to positive cooperativity. Computational analysis predicts subtype specificity of 11-2 F among PRMTs. Structural analysis of ligands in the binding pockets is performed to compare poses of 11-2 F and its redesigned analogs and identifies three new analogs predicted to have significantly better potency. One of them, after synthesis, is ~4 fold more efficient in inhibiting PRMT5 catalysis than 11-2 F, with strong MTA-synergy. These data suggest the feasibility of employing near-atomic resolution cryo-EM structures and computational analysis of ligand poses for small molecule therapeutics. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8cyi.cif.gz 8cyi.cif.gz | 315.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8cyi.ent.gz pdb8cyi.ent.gz | 254.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8cyi.json.gz 8cyi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cy/8cyi https://data.pdbj.org/pub/pdb/validation_reports/cy/8cyi ftp://data.pdbj.org/pub/pdb/validation_reports/cy/8cyi ftp://data.pdbj.org/pub/pdb/validation_reports/cy/8cyi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  27078MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 72766.664 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PRMT5, HRMT1L5, IBP72, JBP1, SKB1 / Production host: Homo sapiens (human) / Gene: PRMT5, HRMT1L5, IBP72, JBP1, SKB1 / Production host:  References: UniProt: O14744, type II protein arginine methyltransferase |
|---|---|
| #2: Protein | Mass: 33226.211 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: WDR77, MEP50, WD45, HKMT1069, Nbla10071 / Production host: Homo sapiens (human) / Gene: WDR77, MEP50, WD45, HKMT1069, Nbla10071 / Production host:  |
| #3: Chemical | ChemComp-P2R / |
| #4: Chemical | ChemComp-MTA / |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Complex of PRMT5 and MEP50 in the presence of MTA and a novel inhibitor (11-2F) Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: .440 MDa / Experimental value: YES |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Conc.: 0.4 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: Preclean plasma cleaner / Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R2/2 |
| Vitrification | Instrument: FEI VITROBOT MARK III / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: DARK FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 1500 nm / Cs: 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 40 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 4115 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 866240 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: D2 (2x2 fold dihedral) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.14 Å / Num. of particles: 207392 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 68 / Protocol: OTHER / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Highest resolution: 3.14 Å |
 Movie
Movie Controller
Controller


 PDBj
PDBj